11.2.1.3—
Ease of Renaturation
A more useful criterion than the buoyant density for detecting chloroplast DNA from higher plants is the ease with which it will renature after heat or alkali denaturation. Nuclear DNA renatures only to a slight extent in a few hours,
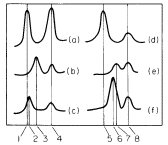
Figure 11.1
Densitometer tracings of ultraviolet absorption photographs of Vicia faba DNA banded
in caesium chloride gradients. (a) Native chloroplast DNA; (b) heat-denatured chloroplast
DNA; (c) renatured chloroplast DNA; (d) native nuclear DNA; (e) heat-denatured nuclear
DNA; (f) renatured nuclear DNA. The numbers refer to densities in g cm-3 as follows: (1)
1.696; (2) 1.699; (3) 1.712; (4) 1.728; (5) 1.696; (6) 1.709; (7) 1.712; (8) 1.728. The
peaks at 1.728 g cm–3 represent marker DNA from Micrococcus radiodurans.
(From Kung & Williams, 1968, by courtesy of Elsevier.)
depending on its content of reiterated sequences. Figure 11.1 illustrates this difference in the case of Vicia faba. The rapid renaturation of chloroplast DNA has encouraged attempts to estimate its genome size from measurements of kinetic complexity. The kinetic complexity of a DNA sample is a measure of the size of the unique set of nucleotide sequences it contains, as judged from the rate at which the DNA renatures. A rapid rate of renaturation implies that like sequences are present in high concentration, and therefore the number of unique sequences, or kinetic complexity, is small. Table 11.2 lists the kinetic complexity, in terms of molecular weight, for the chloroplast DNA from some algae and higher plant species. Some of these values have been corrected from the published figures since the estimate of the molecular weight of the bacteriophage T4 DNA, used as standard, has been revised from 1.3 × 108 to 1.06 × 108 (Dubin et al., 1970). It is striking that the corrected values for the kinetic complexity of chloroplast DNA from the few algae and higher plants examined so far are all in the range 0.9–1.0 × 108 . This may be a coincidence, or it could mean the information content of chloroplast DNA is basically similar throughout the plant kingdom. All the species so far examined belong to the Chlorophyta, with the exception of Euglena. It would be interesting to make such measurements of chloroplast DNA from other plant groups, especially from algae with unusually shaped chloroplasts. Table 11.2 also lists the analytical complexities of chloroplast DNA, i.e. the amount of DNA per chloroplast. Since the kinetic complexities are always much less than the analytical complexities, there must be between 20 and 60 copies of the DNA sequences in each chloroplast. These renaturation studies cannot rule out the possibility of microheterogeneity in nucleotide sequence between the copies but, if it does exist, such heterogeneity is beyond detection by current techniques.
| ||||||||||||||||||||||||||||||||||||