9.7.2—
Ribosomal RNA Synthesis
9.7.2.1—
The Ribosomal RNA Precursor
The DNA that codes for rRNA is known to be associated with nucleoli and the transcription of nucleolar DNA has been studied with the electron microscope (Fig. 9. 11). More detailed information has been obtained by the labelling of intact seedlings, plant segments, or cultured cells, with [32 P]-phosphate or [3 H]-uridine for various times followed by the extraction and characterization of the rapidly-labelled RNA. The mechanism of rRNA synthesis is fairly well understood and a generalized scheme outlining the main features of the process is given in Fig. 9.14. The first stable gene product which can be detected is an rRNA precursor molecule. The molecular weight of this RNA varies in different plants but is generally within the range 2.2–2.6 × 106 daltons. The rRNA precursor from mung bean roots, detected by polyacrylamide gel electrophoresis,
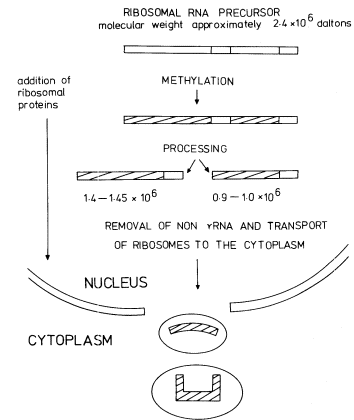
Figure 9.14
The synthesis and processing of the precursor to rRNA.
(Modified from Grierson et al., 1975.)
is shown in Fig. 9.15. The precursor undergoes considerable post-transcriptional modification in the nucleolus before it is converted into 'mature' rRNA and enters the cytoplasm. Shortly after synthesis the molecule becomes methylated at selected sites. This can be demonstrated by incubating plants in methyl-labelled methionine for a short time. The methyl groups become incorporated into the rRNA precursor via s-adenosyl methionine (Cox & Thurnock, 1973). Ribosomal-RNA is known to contain 2'-methyl ribose at certain positions. Only a small number of the residues are modified in this way and it is probable that methylation is completed during the rRNA precursor stage.
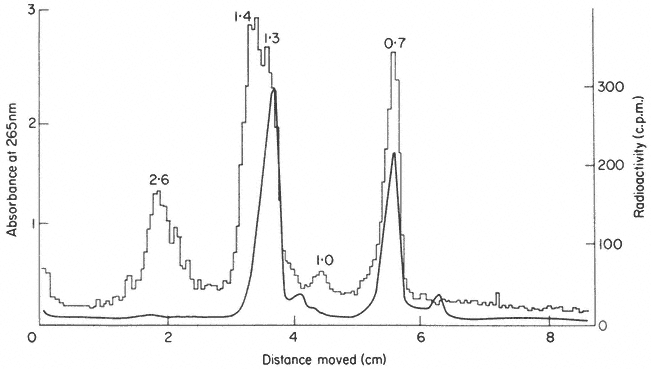
Figure 9.15
Gel electrophoresis of total RNA from roots of P. aureus labelled for
1.5 h with [32 P]-phosphate. The molecular weights of the rRNA precursor and
processing intermediates are shown in millions. (Unpublished result of D. Grierson.)
The precursor molecules correspond to the lateral fibrils attached to the matrix units of active nucleolar DNA (Fig. 9.11). Each molecule contains one sequence of 25s rRNA, one sequence of 18s (total molecular weight = 2.0 × 106 daltons) together with some non-rRNA. The evidence for this is as follows. Firstly, although it is rapidly labelled, the precursor does not accumulate in large amounts. This suggests, therefore, that it must either be rapidly degraded or converted to some other molecular form (Rogers et al., 1970; Leaver & Key, 1970; Grierson et al., 1970; Cox & Turnock, 1973; Grierson & Loening, 1974). This latter explanation would account for the fact that radioactivity appears in mature rRNA slightly later than in the precursor. Secondly, the nucleotide composition of the rRNA-precursor is similar to that of rRNA (Rogers et al., 1970; Leaver & Key, 1970; Grierson & Loening, 1974). Thirdly, competition hybridization experiments have shown that both unlabelled 25s and 18s rRNA compete with radioactive precursor-RNA for sites in the DNA, suggesting that
they share common sequences (Fig. 9.16). Finally, partial digestion of radioactive 25S, 18s and precursor RNA by T1 ribonuclease, followed by two-dimensional fractionation of the fragments shows that the 'fingerprint' pattern of the precursor is similar to that expected of a mixture of 25s and 18s rRNA (Sen et al., 1975).
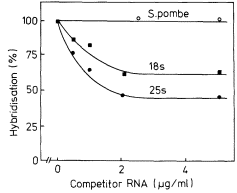
Figure 9.16
Competition hybridization between unlabelled
rRNA and radioactive rRNA precursor for
similar sites in P. aureus DNA.
(From Grierson & Loening, 1974.)
In some plants more than one precursor is detected (Leaver & Key, 1970; Cox & Turnock, 1973). The two precursors from cultured sycamore cells are shown in Fig. 9.17. In some instances the second precursor may originate in chloroplasts or mitochondria (Grierson & Loening, 1974; Kuriyama & Luck, 1973), but in carrot and sycamore both precursors are thought to be in the nucleus (Leaver & Key, 1970; Cox & Turnock, 1974). It is not clear in these cases whether the two RNAs contain similar sequences or whether they are transcribed from different genes. It is possible that the larger RNA is converted to the smaller one, but careful analysis of the rate of labelling of both molecules does not support this suggestion (Cox & Turnock, 1973; Leaver & Key, 1970).
9.7.2.2—
Processing of the Precursor
In addition to becoming methylated, and while still in the nucleolus, the precursor becomes associated with protein molecules and is processed by nucleolytic enzymes which remove the excess non-rRNA in stages to produce mature rRNA in ribosomal subunits ready for transport to the cytoplasm. Some RNA processing intermediates are present in the sample shown in Fig. 9.15. The scheme for processing outlined in Fig. 9.14 is consistent with some, but not all, the studies on rRNA synthesis in plants. For example, no immediate precursor to 25s rRNA is detectable in rye embryos (Sen et al., 1975). Furthermore the 1.0 × 106 daltons RNA is not always detected wheras certain additional RNAs
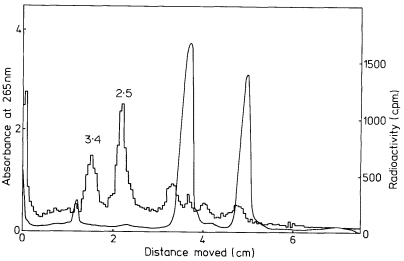
Figure 9.17
Pulse-labelled RNA from sycamore cells showing two rRNA precursor
molecules. The cells were labelled for 40 minutes with [3 H]-uridine and
total nucleic acid fractionated by gel electrophoresis. The DNA was not
removed but an identical distribution of radioactivity is observed when
this is carried out. The approximate molecular weights of the precursor
RNAs are indicated in millions. (Unpublished result of D. Grierson.)
are sometimes observed. For example, after cycloheximide treatment of cultured parsley cells, which slows down rRNA processing, previously undetected RNAs with molecular weights of 2.0 and 0.9 × 106 daltons were found (Gebauer et al., 1975). This suggests that if certain processing events occur very rapidly they may normally go undetected. It is very probable that the complete sequence of processing steps will only be worked out when it becomes possible to study accurately RNA metabolism in isolated nucleoli. Apparent differences between closely related species may be explained by assuming that certain critical stages of processing occur at different rates. In addition, it should be realised that the processing enzymes are functioning at different, possibly distant, sites within the same molecule. There seems no compelling reason to expect these modifications always to occur in the same chronological order and, if they do not, variations in the pattern of processing intermediates might be expected under altered conditions or in different species.
The function of the excess RNA in the precursor is not known. It is too variable in size in different organisms to be a mRNA for the ribosomal proteins (Loening, 1970) and in any case it appears to be degraded very rapidly after synthesis. It may play some role in the regulation of transcription but more probably it is necessary for ensuring that rRNA adopts the correct secondary and tertiary structure required for methylation and for the addition of proteins during ribosome assembly.
It is pertinent to ask whether the multi-stage mechanism that operates for rRNA production is unique or whether it represents a general principle common to the synthesis of all types of RNA. Precursors to tRNA and 5s RNA have been identified and characterized in bacteria and animals (Hecht et al., 1968; Bernhardt & Darnell, 1969) and precursors of mRNA have also been detected, although there is uncertainty about the actual details of processing (Brawerman, 1974). Although RNA metabolism has not been so intensively studied in plants the indications are that precursors to 5s RNA, tRNA and mRNA do occur. When considering the question of the regulation of RNA metabolism, therefore, it is necessary to take account of post-transcriptional modifications as possible control points.