9.4.5—
Repeated Sequence DNA
Information about the sequence content of DNA can be obtained by studying the renaturation rate. Double-stranded DNA is first sheared to lengths of a few hundred nucleotides by ultrasonication or by forcing through a narrow gauge needle. This is designed to liberate short sequences of DNA and to allow them subsequently to react independently of neighbouring sequences. The DNA is then denatured and allowed to renature at the desired concentration under controlled conditions. Renaturation is followed by monitoring the absorbance of the solution at 260 nm in a spectrophotometer cell maintained at the appropriate temperature, or alternatively by taking samples at intervals and fractionating them by hydroxyapatite chromatography. In some situations partially double-stranded segments of DNA are produced, which have single-stranded 'tails'. These can be removed by treating the reassociated DNA with SI nuclease, which is specific for single-stranded DNA, before hydroxyapatite fractionation.
The reassociation reaction follows second order kinetics, which means that it is governed by the concentration of the reacting sequences. This is determined by the DNA concentration and the sequence content. For example, synthetic poly-dA. dT renatures almost instantaneously, whereas at the sme concentration naturally occurring DNA takes much longer to renature. In general the renaturation rate is inversely proportional to the number of base pairs in the genome (providing that no repetitive sequences are present). In principle, therefore, the number of 'genes' or sequences may be estimated by comparing the renaturation rate of an unknown DNA with that from an organism with a well characterized genome. The results of renaturation experiments are often expressed as a 'Cot' plot. In this form the extent of DNA renaturation is plotted as a function of the initial concentration of the DNA (in moles of nucleotides per litre) multiplied by the renaturation time (in seconds) plotted on a log scale. One advantage of this is that different types of DNA can easily be compared by their Cot 1/2 values (the value of Cot at which 50% of the DNA becomes renatured) (Fig. 9.7a). For a variety of microorganisms and viruses the Cot 1/2 is related to the genome size. However, this is not strictly true for eukaryotic DNA which behaves in a more complex way.
In bacteria and viruses most DNA sequences are represented only once in the genome but in higher organisms many sequences occur in multiple copies. For this reason a plot of the renaturation of higher plant DNA deviates markedly
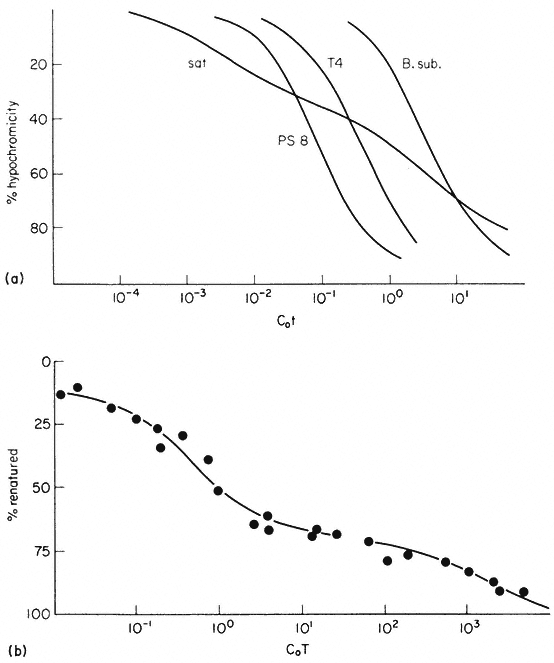
Figure 9.7
Reassociation kinetics of various DNA samples. (a) Musk melon satellite
(sat.), phage PS8 (PS8), Bacillus subtilis (B. sub. ), phage T4 (T4). The data
are corrected for differences in GC content and to take account of different
solvent concentrations. (b) Helianthus annuus DNA. The slowest reassociating
fraction behaves approximately as expected for sequences present in one copy per
haploid genome. (a) redrawn from Bendich and Anderson, 1974; (b) from Flavell et al., 1974.)
from a second order curve (Fig. 9.7b). A substantial proportion of the genome is so highly repetitive that it renatures even more rapidly than bacterial DNA. A second, intermediate, fraction contains families of repetitive sequences present in a lower frequency. Finally, there exists a unique fraction of DNA which essentially comprises sequences present in only one or a small number of copies in the genome. Flavell et al., (1974) have studied the reassociation kinetics of the DNA from 23 higher plant species, which vary in 2C nuclear DNA content between 1.7 and 98 × 10–12 g. The results, summarized in Table 9.4, show that in plants with relatively large genomes an average of 80% of the DNA consists of sequences present in 100 copies or more. For plants with smaller genomes
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
the average amount of repetitve DNA is 62%. These results indicate that part of the variation in nuclear DNA mass can be accounted for by variation in the amount of repeated sequence DNA.