SECTION TWO—
GENE EXPRESSION AND ITS REGULATION IN PLANT CELLS
Introduction
The activities, potentialities, and developmental fate of all cells are determined by the hereditary genetic material stored within the nucleus, and also to a lesser but still important extent, by that stored within the plastids and mitochondria. Hereditary information is coded within the nucleotide sequences of the DNA, and the generalized concept of the expression of genetic information is usually portrayed as follows:
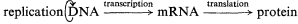
This 'central dogma' states that the information contained within the DNA is only expressed, in terms of cellular processes, through the action of the ultimate gene products, the proteins, most of which are enzymatic in nature. The central dogma also points to there being two principal points at which gene expression may be regulated; i.e. gene transcription and m-RNA translation. In fact, a further control point should be added, since the catalytic activity of enzymes may be regulated in several ways in addition to the regulation of the synthesis of the enzyme. Thus the regulation of gene expression may be more adequately described by the following scheme:
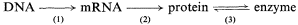
in which the potential control points at transcription, translation and enzyme activation/inactivation are numbered.
Section II considers the biochemical and structural basis of gene expression and its regulation in higher plants and places it in context with what is known from the more extensive studies carried out with other organisms. The principal hypothesis of the regulation of gene expression stems from the discoveries relating to the control of enzyme levels in bacteria. In 1961, to account for the known effects of metabolites on the synthesis of specific enzymes in bacteria, Jacob and Monod proposed that the activity of the genes which code for specific enzyme proteins (structural genes ) is controlled by other genes, not necessarily linked to the structural genes, and known as regulator genes. A regulator gene codes for the synthesis of a specific cytoplasmic protein, known as a repressor, which has the capacity to interact with a region of chromosomal DNA (the operator ) adjacent to the initiation end of the structural gene. The binding of a repressor to a specific operator, which may control a sequence of structural genes (known collectively with the operator as an operon ) inhibits transcription of m-RNA on those structural genes. It is also held that the repressor protein has the capacity to bind one or more low molecular weight
substances, known as effectors, which may alter its capacity to bind to the operator. Effectors which combine with the repressor and prevent its binding to the operator are termed inducers, since they allow enzyme synthesis from a previous repressed state; inducers are commonly substrates of the first enzyme in a pathway. Effectors which combine with the repressor and enhance binding with the operator, inhibit or repress enzyme synthesis; such effectors are known as co-repressors and are commonly products of a reaction pathway. In repressible systems of this nature the structural genes are normally fully expressed in the absence of the co-repressor and the enzymes are thus known as constitutive. As shown in Fig. 11.1 this hypothesis elegantly accounts for the formation of enzyme in response to the substrate (enzyme induction ) and for the cessation of enzyme formation in response to the end product (enzyme repression ).
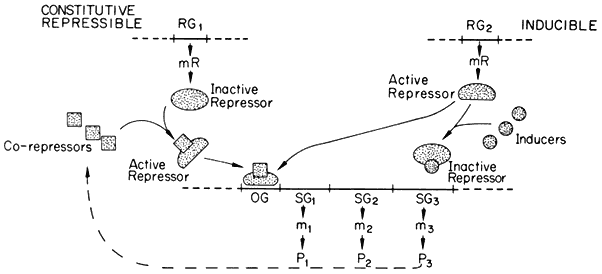
Figure 11.1
A generalized scheme of the Jacob–Monod model for constitutive repressible
and inducible systems of gene regulation. The regulator genes (RG) are shown as
coding for the formation of either active repressors (Inducible system) or inactive
repressors (Constitutive repressible system) which bind to the operator gene (OG)
and block the transcription of the structural genes of that operon (SG1 to SG3 ). P =
protein and m = mRNA; P3 is considered to be an enzyme whose catalytic activity
leads to the formation of a co-repressor for the constitutive repressible system.
(Modified with permission from F.A.L. Clowes and B.E. Juniper
(1968) Plant Cells. Blackwell Scientific Publications Ltd., Oxford.)
It is not yet clear how fully these concepts account for gene expression and its regulation in eucaryotic organisms. As will be seen from Chapter 9, the structure, organization and complexity of nuclear DNA are wholly different from the rather simple arrangements found in bacteria. There seems to be far too much DNA than would be necessary to code for the predicted number of structural genes and attendant regulator genes as estimated from the Jacob-Monod model. Several suggestions have been put forward to account for this unexpected genetic largesse, but the most likely ideas involve the extra DNA in the regulation of the structural genes (see Britten & Davidson, 1969).
Additionally, the separation of the site of transcription (in the nucleus) from that of translation (in the cytoplasm) creates further opportunities for regulation. It will be seen that the processes of RNA and protein synthesis in the nuclei and cytoplasm of plants are similar in principle, but different in detail, from those in higher animals (chapters 9 and 10).
A very important aspect of plant molecular biology, and one which is unique to plants, is the presence of a substantial store of genetic information in the chloroplasts (chapter 11). The coding potential of chloroplast DNA is vastly greater than that of mitochondrial DNA, and many of the activities of higher plant cells, and indeed a large part of their total protein, is coded for in the chloroplast genome. The regulatory processes involved here must be highly complex, since it is known that some chloroplast proteins are coded for and synthesized within the plastids, and others coded in the nucleus, and synthesized in the cytoplasm; Fraction I protein, the most abundant protein in green leaves, is composed of two types of sub-units, one of nuclear, and one of plastid, origin. Clearly, here the regulation of gene expression must involve the integration of transcription and translation within three separate cell compartments, the nucleus, the cytoplasm and the chloroplast.
As stated briefly above, gene expression may also be regulated after the final gene product, the protein, has been formed, since a gene can only be considered to be expressed when the protein it codes for performs its specific function. Thus the activation and inactivation of enzymes, through the several known mechanisms, becomes properly a facet of gene expression (chapter 12).
The integration of the activities of different cells, particularly with regard to development, inevitably requires on the one hand, the generation and transmission of intra- and inter-cellular stimuli and, on the other, specific responses to these stimuli. In higher animals, both electrical and chemical stimuli appear to be important; in higher plants inter-cellular integration seems to be almost completely chemical, through a narrow range of hormonal substances. Although understanding of the mechanism of hormone action in plants is still incomplete, the regulation of gene expression is probably involved (chapter 13).
All organisms need to be able to perceive their environment and adapt to environmental changes. Evolution, having favoured non-motility for plants, has inevitably endowed them with extremely subtle mechanisms for detecting and responding to certain environmental factors. The best example here is the response of plants to light, in which specific photoreceptors react to signals from the radiation environment by initiating changes in metabolism and development. As an example of external regulation of gene expression in higher plants, chapter 14 deals with the mechanism of action of phytochrome, an important photoreceptor found in all green plants.
Section II therefore, is concerned with the genetic material of higher plants, its organization, complexity and regulation, with particular reference to the control of cell metabolism, to cell-cell communication in hormone action and to responses to the environment via photoreceptor action.
Chapter 9—
The Nucleus and the Organization and Transcription of Nuclear DNA
9.1—
Introduction
The importance of the cell nucleus has been recognized for some considerable time and a great deal of literature concerned with its general organization and composition and the structure and behaviour of chromosomes has been derived from cytological investigations. This present chapter is mainly concerned with recent developments in research which provide a more detailed understanding of the organization and function of nuclear DNA, the mechanism of RNA synthesis and the question of the control of gene expression in higher plants.
9.2—
Nuclear Structure and Composition
In general, nuclei are spherical or disc-shaped, although occasionally they may appear lobed. They vary in size in different species but are usually within the range 1–10 µm in diameter (Fig. 9.1). Nuclei may be isolated by gently breaking cells in a suitable osmoticum, filtering the homogenate, for example through muslin, followed by low-speed centrifugation. Attempts are usually made to free the nuclei of contaminating cytoplasm, cell wall fragments and other organelles by centrifugation through sucrose. However, methods for obtaining high yields of intact, purified nuclei from higher plant cells are at present not very satisfactory. For certain purposes it is often easier to work with chromatin preparations which consist essentially of chromosomal and nuclear material obtained from broken nuclei.
The major part of the cellular DNA is present in the nucleus, arranged in a highly organized fashion within the chromosomes which at different times may show characteristic bands or loops (see Fig. 9.1). Chromosomes are composed of approximately equal amounts of DNA and basic proteins known as histones (see Table 9.7). In addition, much smaller quantities of RNA and acidic proteins are present which are probably of great importance in controlling gene expression. There is wide variation in the number of chromosomes observed within the nuclei of different species although in diploid cells of a single species the number is normally constant. During interphase the chromosomes are less highly condensed than at mitosis and are more difficult to detect. At this stage at least part of the DNA is being transcribed into RNA. It is possible to identify chromosomes on the basis of morphological features such as size, position of the centromere and the presence or absence of constrictions. In addition, certain
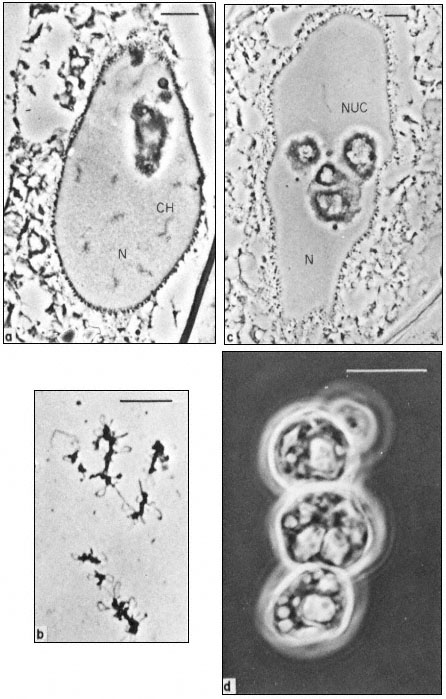
Figure 9.1
Phase contrast light micrograph of nuclei and isolated nuclear
contents from Acetabularia. These giant nuclei are unusually large;
the scale line represents 10 µm. Chromosomes (CH) and nucleoli (NUC)
can be seen within the nuclei (N) fixed in situ (a and c) and the appearance
of isolated chromosomes and nucleoli are shown in b and d. Note the large
aggregates of nucleolar material with an internal cavern system (c and d) and the
'lampbrush-chromosome-like' morphology of the chromosomes (b). (Fig. 9.1a, c and d
from Spring et al., 1974; 9.1b, previously unpublished micrograph from the same authors.)
areas of heterochromatin, which appear more highly condensed and therefore take up more stain, may be distinguished from the remainder of the chromosome, termed euchromatin. In certain cell types, such as embryo suspensor cells, giant polytene chromosomes occur. In such situations it is possible to study the structural organisation of chromosomes in much greater detail.
Recent developments have made it possible to identify individually each of the 46 chromosomes in man on the basis of characteristic banding patterns produced by certain staining procedures. These techniques and their application to the study of plant chromosomes have been reviewed by Vosa (1975).
Organelles known as nucleoli become visible within the nucleus during interphase. These are very prominent spherical bodies with different staining properties to the remaining part of the nucleus. They arise in association with specific regions, the nucleolar organizers, of certain chromosomes. Nucleoli are not surrounded by a membrane but they do have a highly ordered structure (Fig. 9.2). The central core consists of a region of fibrils 8–10 nm across surrounded by a region of dense granules 15–20 nm in diameter. Mutants which lack nucleoli are unable to synthesise ribosomes and it is now known that the genes coding for ribosomal RNA normally occur at the nucleolar organizer region of chromosomes. Here the DNA is transcribed into ribosomal-RNA (rRNA) which is assembled to form ribosomes in the outer region of the nucleolus by combination with ribosomal proteins. The nucleolus is most prominent, therefore, when synthesising large numbers of ribosomes. This explains the old observation that there is a general correlation between nucleolar volume and the rate of protein synthesis of cells.
The fact that nuclei are surrounded by a membrane was deduced from the observation that they are selectively permeable to small molecules. The nature of the double membrane may be readily observed with the aid of the electron microscope (Roberts & Northcote, 1971). It is often interconnected with the cytoplasmic membrane system, the endoplasmic reticulum, and is perforated by a large number of pores. This is demonstrated quite clearly in Fig. 9.3 which shows a surface view of a portion of the nuclear membrane of Acetabularia. The apparent diameter of a nuclear pore is of the order of 100 nm. However, the observation that colloidal particles of much smaller size do not readily pass through these pores suggests that their effective size is substantially less, probably due to the presence of a protein-containing matrix within the pore.
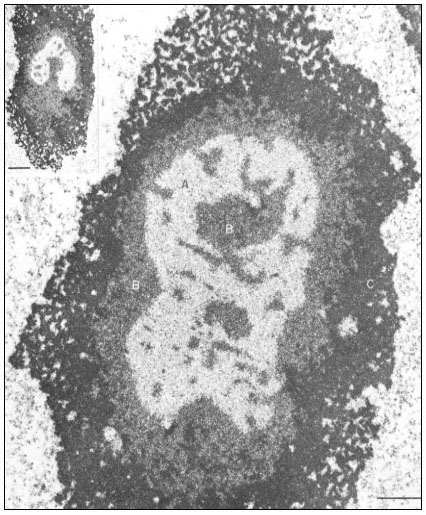
Figure 9.2
Ultrastructural details of the composition of the nucleolar subunits of Acetabularia
revealed by electron microscopy. Note the finely fibrillar texture in the internal zone
A and the dense packing of small granulofibrillar structures in zone B, extensions
from which deeply penetrate zone A (see also the insert ) and the very dense
packing of granules in the cortical zone C. The scale indicates 1 m m,
the scale in the insert denotes 2.5 µm. (From Spring et al., 1974.)
9.3—
Nuclear DNA Content
The amount of DNA present in nuclei may be estimated by nuclear isolation followed by extraction and direct chemical measurement of DNA content. A more elegant approach is to carry out quantitative microspectrophotometry of
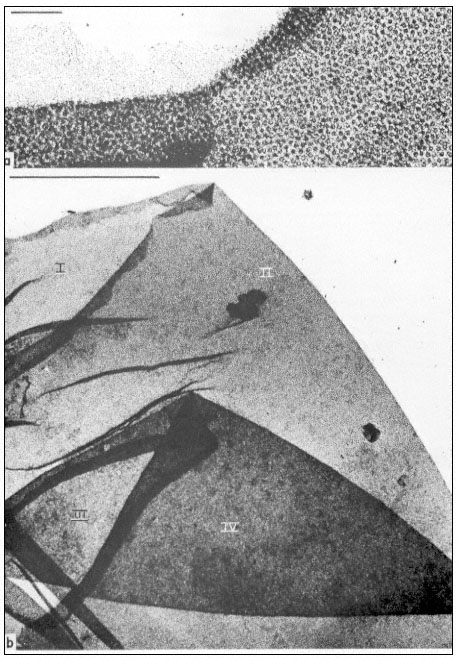
Figure 9.3
Manually isolated nuclear envelope of Acetabularia mediterranea. This shows
a sequence of overfoldings (I to IV in b). Note the high density of nuclear pore
complexes and the association with some fibrillar material as identified in the
upper region of a. The scale in (a) denotes 1 µm, the scale in (b) denotes 10 µ m.
(From Spring et al., 1974.)
individual nuclei following Feulgen staining. Table 9.1 compares the nuclear DNA content of a range of higher plants with the amounts of DNA found in lower organisms. Two conclusions can be drawn from such a comparison. Firstly, and perhaps not surprisingly, higher plants contain considerably more DNA than bacteria and fungi. For example, Pisum sativum contains 1,000 times as much DNA as Escherischia coli . Secondly, however, the amount of DNA present in different higher plants may vary by as much as 100 fold. Further comparison shows that there is little correlation between chromosome number and DNA content.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||
One explanation of the increased DNA content of higher plants is that more complex organisms require a greater store of genetic material than bacteria and viruses. Although it is difficult to estimate precisely the extent of the increased coding requirement it is quite clear that this is in itself not a satisfactory explanation for the presence of such large amounts of DNA in some plants. There often exists a wide variation in the 2 C DNA content of plant species within one: genus and it is unreasonable to conclude that small differences between related plants are a direct consequence of gross differences in nuclear DNA. Furthermore, some plants contain 20 times as much DNA as mammals and yet the latter are normally accepted as comprising a more complex group of organisms. It therefore appears that the amount of nuclear DNA is not directly related to the complexity of the organism. It is also possible to argue on other grounds that the total amount of nuclear DNA is not required in order to code for additional proteins. Firstly, it is difficult to envisage functions to account for a 1,000 fold increase in the number of enzymes. Secondly, if each higher plant contained such a large number of essential genes it would, on numerical grounds, be prone to an unrealistically high incidence of mutation.
The size of the genome of plants with relatively low nuclear DNA contents is of the order of 2 × 1012 daltons. Given that the molecular weight of a gene required to code for a messenger-RNA (mRNA) molecule for an average-sized polypeptide is 106 daltons, this represents, in principle, 2 × 106 genes. A more realistic estimate would be that 1% of this DNA actually codes for proteins. Experimental evidence for this view and a discussion of the organization, properties and possible function of the remaining DNA is given later.
It used to be widely proclaimed that when differences in ploidy are taken into account the amount of DNA in each nucleus of a single species is constant. The observation that the gene number remains constant, and the fact that many plant cells are totipotent, supports the view that differentiation involves the selective control of gene expression. However, the rule of DNA constancy is not inviolate. It is now known that specific regions of the genome may become selectively amplified at certain stages of development. This undoubtedly occurs to the genes for ribosomal RNA during oocyte development in Amphibia. Although there exist no established cases of ribosomal RNA gene amplification in higher plants, the satellite DNA present in melon may be under- or over-replicated in different tissues (Ingle & Timmis, 1975).
One very intriguing observation is that the amount of nuclear DNA in flax may become altered if the plants are subjected to certain regimes of nitrogen and phosphorus fertilizer. The alteration in nuclear DNA content is stable and inherited (Evans, 1968). Although this phenomenon is well established it is not known how widespread it may be and there is no satisfactory explanation to account for it.
9.4—
Fractionation and Properties of DNA
9.4.1—
General Properties
Although single-stranded DNA does occur naturally most DNA molecules consist of two anti-parallel chains of nucleotides. Each nucleotide consists of deoxyribose, a phosphate group and a base. The four bases that occur in all types of DNA are the purines adenine and guanine and the pyrimidines thymine and cytosine. Adenine is capable of forming hydrogen bonds with thymine, and guanine with cytosine. Sometimes modified bases are present in small amounts and in plant nuclear DNA a substantial proportion of the cytosine residues are replaced by 5-methyl-cytosine. In double stranded DNA there is an equivalence between the amount of adenine and thymine and guanine and cytosine plus 5-methyl-cytosine. The two strands are joined together by hydrogen bonds between complementary pairs of bases to form a right handed helix. The bases are at right angles to the sugar residues, and project inwards, and the phosphate groups are on the outside. In the B form of DNA there are ten base pairs in one complete turn of the helix, which extend for 3.4 nm. The width of the helix is 1.8 nm. An alternative configuration is possible, the A form, where the bases are tilted and 11 pairs are accommodated per turn of the helix. Double stranded RNA and DNA-RNA hybrids adopt this latter configuration.
Many DNA molecules are known to be several millimetres in length. It is very difficult to determine the actual length of very large DNA molecules because they are extremely sensitive to shearing forces produced by various isolation procedures. It is known that the complete genome of E. coli, with a molecular
weight of 2.9 × 109 daltons, consists of a single circular DNA molecule of length approximately 1 mM . The total amount of DNA in a higher plant cell is equivalent to a length of approximately 1 m, but it is not known how many molecules this comprises.
The base composition of DNA is normally expressed as the per cent G + C content. This value, which is characteristic of a given species, varies widely in different organisms and in bacteria may be anything from 30% to 70%. The base compositions of a number of plant DNAs are given in Table 9.2.
9.4.2—
Buoyant Density Centrifugation
One of the most common methods of fractionating DNA is to exploit differences in density between different types of DNA in concentrated salt solutions. Caesium chloride is the most commonly used. The method involves prolonged high speed centrifugation of purified DNA in concentrated caesium chloride. The centrifugation produces a gradient of salt concentration in the centrifuge tube and the DNA migrates to occupy a position in the gradient which corresponds to its own buoyant density. A number of factors affect the buoyant density, such as the nature of the caesium salt, the presence of heavy metals or DNA-binding dyes, the pH and the temperature. Under constant conditions (usually 25°C in caesium chloride at neutral pH) the buoyant density of DNA is related to the GC content:
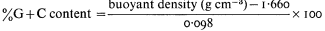
DNAs with different base compositions can therefore be separated by this method. Figure 9.4 shows the fractionation of Phaseolus aureus DNA (buoyant density = 1.695) and E. coli DNA (buoyant density = 1.710) in caesium chloride.
Most nuclear DNAs from higher plants have buoyant densities within the range 1.69–1.71 g cm–3 . However, the presence of 5-methyl-cytosine serves to reduce the density slightly, thereby giving rise to an under-estimate of the GC content. In general, 1% methylation decreases the buoyant density by 1 mg cm–3 . Certain sequences of bases may also distort the relationship between base composition and buoyant density. Furthermore, single-stranded DNA is denser than double-stranded DNA of similar base composition by approximately 0.015 g cm–3 and under alkaline conditions the density is increased by 0.06 g cm–3 . This is due partly to the fact that DNA becomes single-stranded under these conditions and also partly because the deprotonated adenine and thymine residues are neutralized by binding caesium ions.
9.4.3—
Satellite DNA
Figure 9.4 shows that DNA from a particular organism often forms an apparently homogenous band in caesium chloride. The width of the band depends
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
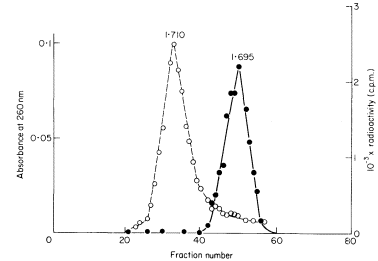
Figure 9.4
The separation of a mixture of E. coli and P. aureus DNA in caesium chloride.
A trace of E. coli DNA labelled with 3 H thymidine was mixed with P. aureus DNA
and centrifuged at 30,000 rpm at 25ºC in caesium chloride for 3.5 d using a fixed-angle
centrifuge rotor. Three-drop fractions were collected and the P. aureus DNA located
by absorbance at 260 nm (solid circles, continuous curve) and E. coli DNA detected by
measurement of radioactivity (open circles, dotted curve). (Unpublished result of D. Grierson.)
upon the slope of the density gradient, the molecular weight of the DNA and the difference in base composition between different regions of the DNA. In most plants certain regions of the genome are sufficiently different in base composition for them to occupy a position distinct from the main-band DNA. If they represent only a small percentage of the total DNA such sequences are difficult to detect but when present in large amounts they produce one or more peaks of satellite DNA. Melon DNA provides an example where the satellite is denser than the main-band (Fig. 9.5) but light satellites have also been observed, for example in flax. It has been assumed that the majority of satellite DNAs are nuclear in origin but recent studies with cucumber suggest that they may sometimes represent organelle DNA (Kadouri et al., 1975). Although they are widely distributed in plants no satellites have so far been found in monocotyledonous plants using caesium chloride fractionation (Table 9.3). However similar components can be detected using other techniques. For example, substances such as heavy metals and dyes often selectively bind to certain sequences within the DNA. This alters their buoyant density and thus generates satellites. One common method is to fractionate DNA in caesium sulphate gradients in the presence of silver ions. Using this procedure, Ingle (unpublished) has demonstrated the presence of a satellite DNA in Scilla (a monocotyledon) although in caesium chloride no such satellite is observed.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
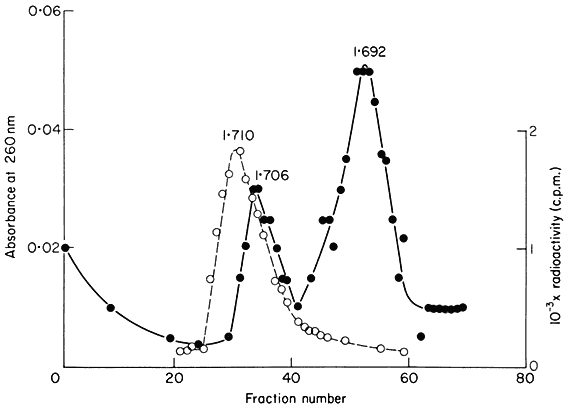
Figure 9.5
The separation of a mixture of E. coli and melon DNA in caesium chloride.
The experimental details were as described in the legend to Fig. 9.4. Melon
DNA was detected by absorbance at 260 nm (solid circles, continuous curve)
and E. coli DNA by measurement of radioactivity (open circles, dotted curve).
(Unpublished result of D. Grierson.)
9.4.4—
Dissociation and Reassociation of DNA
When double-stranded DNA in solution is heated above a certain temperature the hydrogen bonds between complementary base pairs are broken and the two strands dissociate. The process, which can also be brought about by treatment of DNA with alkali, is commonly referred to as melting or denaturation. Strand separation is accompanied by an increase of approximately 35% in the absorbance of ultra-violet light. This hyperchromic effect provides a means of monitoring the dissociation of double-stranded DNA by measuring the increase in absorbance of a DNA solution at 260 nm as the temperature is raised. The melting curves of DNA from a variety of sources are compared in Fig. 9.6. The temperature at which 50% of the DNA is melted, the Tm, depends upon a number of factors such as the salt concentration and pH. Under standard conditions, i.e. when the DNA is dissolved in standard saline citrate (S.S.C. being 0.15 M sodium chloride, 0.015 M trisodium citrate, pH 7) the base composition and the Tm are related in the following way:
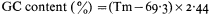
In viruses, the majority of sequences within the genome are similar in base composition and for a given DNA a sharp melting profile is observed with a characteristic Tm. In contrast, higher organism DNA often consists of distinct subfractions, each with a slightly different Tm. This produces a broad melting
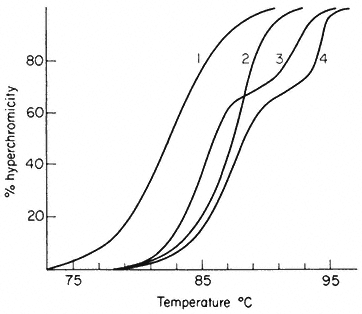
Figure 9.6
Melting curves of various DNA samples. (1) Musk melon main-
band and (4) satellite DNA. (3) a 2:1 mixture of DNA from bacterium
C and phage P58. (2) B. subtilis DNA. Melting was carried out in S.S.C.
(From Bendich & Anderson, 1974.)
profile which represents a number of overlapping melting curves. With DNA samples from organisms that contain prominent satellite bands in caesium chloride the melting curve often has two or more phases. Even where no large satellites are present heterogeneity within the DNA can readily be detected by plotting the rate of change of absorbance at 260 nm of a DNA solution as a function of temperature.
If, following denaturation of DNA by alkali or by heating, the pH of the solution is restored to neutrality or the temperature reduced below the Tm, complementary strands of DNA show a marked tendency to recombine to form double-stranded molecules. This process of duplex formation is termed renaturation or reassociation. The rate of the reaction is governed by the concentration of DNA sequences, pH, temperature and salt concentration. Renaturation is normally carried out at moderately high salt concentrations at a pH close to neutrality and at a temperature approximately 25°C below the Tm. Under these conditions, single-stranded DNA molecules collide at random. Most collisions occur in such a way that complementary sequences of bases are not in register and the strands do not reassociate. If, however, two strands approach in an anti-parallel configuration and potential regions of complementarity are in close proximity a nucleation event will take place and hydrogen bonds will form. Whether or not the strands remain together depends upon the nature of the neighbouring bases on either side of the nucleation site. If complementary sequences are arranged in the correct register along the length of the molecule a rapid zippering effect takes place to produce a stable double-stranded DNA molecule.
DNA renaturation can be followed by measuring any property that distinguishes single-stranded from double-stranded DNA. The decrease in absorbance at 260 nm that accompanies duplex formation is often exploited for
this purpose but fractionation by hydroxyapatite is also commonly used. Under appropriate conditions only double-stranded DNA binds to hydroxyapatite (a hydrated form of calcium phosphate); single strands pass straight through a column and double-stranded material may subsequently be eluted either by increasing the salt concentration or by raising the temperature above the Tm. This procedure has the advantage that the properties of the single-stranded and renatured fractions may subsequently be studied separately.
9.4.5—
Repeated Sequence DNA
Information about the sequence content of DNA can be obtained by studying the renaturation rate. Double-stranded DNA is first sheared to lengths of a few hundred nucleotides by ultrasonication or by forcing through a narrow gauge needle. This is designed to liberate short sequences of DNA and to allow them subsequently to react independently of neighbouring sequences. The DNA is then denatured and allowed to renature at the desired concentration under controlled conditions. Renaturation is followed by monitoring the absorbance of the solution at 260 nm in a spectrophotometer cell maintained at the appropriate temperature, or alternatively by taking samples at intervals and fractionating them by hydroxyapatite chromatography. In some situations partially double-stranded segments of DNA are produced, which have single-stranded 'tails'. These can be removed by treating the reassociated DNA with SI nuclease, which is specific for single-stranded DNA, before hydroxyapatite fractionation.
The reassociation reaction follows second order kinetics, which means that it is governed by the concentration of the reacting sequences. This is determined by the DNA concentration and the sequence content. For example, synthetic poly-dA. dT renatures almost instantaneously, whereas at the sme concentration naturally occurring DNA takes much longer to renature. In general the renaturation rate is inversely proportional to the number of base pairs in the genome (providing that no repetitive sequences are present). In principle, therefore, the number of 'genes' or sequences may be estimated by comparing the renaturation rate of an unknown DNA with that from an organism with a well characterized genome. The results of renaturation experiments are often expressed as a 'Cot' plot. In this form the extent of DNA renaturation is plotted as a function of the initial concentration of the DNA (in moles of nucleotides per litre) multiplied by the renaturation time (in seconds) plotted on a log scale. One advantage of this is that different types of DNA can easily be compared by their Cot 1/2 values (the value of Cot at which 50% of the DNA becomes renatured) (Fig. 9.7a). For a variety of microorganisms and viruses the Cot 1/2 is related to the genome size. However, this is not strictly true for eukaryotic DNA which behaves in a more complex way.
In bacteria and viruses most DNA sequences are represented only once in the genome but in higher organisms many sequences occur in multiple copies. For this reason a plot of the renaturation of higher plant DNA deviates markedly
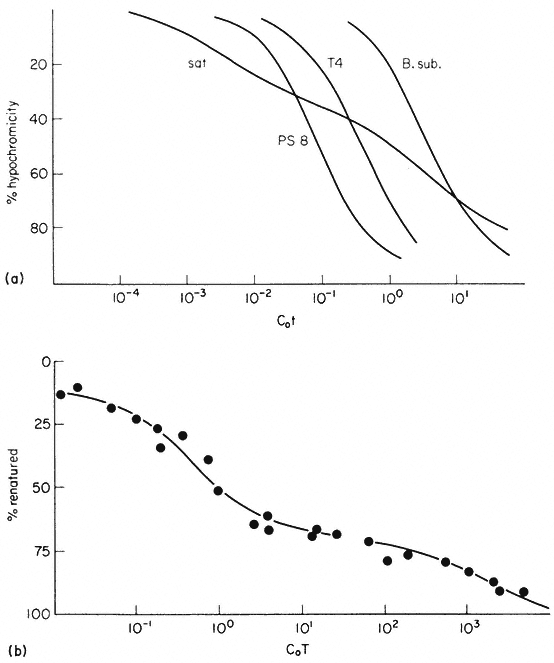
Figure 9.7
Reassociation kinetics of various DNA samples. (a) Musk melon satellite
(sat.), phage PS8 (PS8), Bacillus subtilis (B. sub. ), phage T4 (T4). The data
are corrected for differences in GC content and to take account of different
solvent concentrations. (b) Helianthus annuus DNA. The slowest reassociating
fraction behaves approximately as expected for sequences present in one copy per
haploid genome. (a) redrawn from Bendich and Anderson, 1974; (b) from Flavell et al., 1974.)
from a second order curve (Fig. 9.7b). A substantial proportion of the genome is so highly repetitive that it renatures even more rapidly than bacterial DNA. A second, intermediate, fraction contains families of repetitive sequences present in a lower frequency. Finally, there exists a unique fraction of DNA which essentially comprises sequences present in only one or a small number of copies in the genome. Flavell et al., (1974) have studied the reassociation kinetics of the DNA from 23 higher plant species, which vary in 2C nuclear DNA content between 1.7 and 98 × 10–12 g. The results, summarized in Table 9.4, show that in plants with relatively large genomes an average of 80% of the DNA consists of sequences present in 100 copies or more. For plants with smaller genomes
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
the average amount of repetitve DNA is 62%. These results indicate that part of the variation in nuclear DNA mass can be accounted for by variation in the amount of repeated sequence DNA.
9.5—
The Genes for Ribosomal-RNA
More is known about the properties, organization and function of the genes for rRNA than any other part of the genome. These DNA segments direct the synthesis of rRNA molecules which form the structural 'backbone' of ribosomes, without which cells cannot function. It has been known for some time that they are associated with the nucleolus because certain mutants of the toad Xenopus, which lack nucleoli, are incapable of synthesizing rRNA. During development these mutants die because they are unable to synthezise ribosomes.
9.5.1—
DNA-RNA Hybridization
The genetic origin of any RNA molecule can be studied by DNA-RNA hybridization. This technique is similar in principle to DNA-DNA reassociation. It depends upon the fact that any RNA molecule synthesized on a DNA template has a base sequence complementary to the DNA strand from which it was transcribed. Under suitable conditions, therefore, DNA can be induced to combine with complementary-RNA (cRNA) to produce an RNA-DNA double helix. This provides a means of locating and measuring the genes that code for a particular type of RNA.
One of the commonest procedures is to prepare pure DNA, denature it by heating or treatment with alkali, and to attach the single strands to a solid matrix such as a nitrocellulose filter. During subsequent treatments the immobilized DNA strands are prevented from reassociating but they are capable of forming hybrids when they are placed in a solution of nucleic acid containing complementary base sequences. Using radioactive RNA the formation of DNA-RNA hybrids can be detected by measuring the amount of radioactivity that becomes associated with the filter. The reaction is affected by a number of factors such as RNA concentration, salt concentration, temperature, the presence of compounds such as formamide, which lowers the Tm, and the reaction time. With rRNA at a concentration of 2 µg cm–3 at 70°C in 6 × S.S.C. the reaction is virtually complete within a couple of hours (Fig. 9.8). After a suitable time the filters are washed, given a mild treatment with ribonuclease to remove any unhybridized RNA, and the amount of radioactive RNA remaining is measured in a liquid scintillation counter. If the amount of DNA initially applied to the filter is known then the extent of hybridization can be calculated from the specific radioactivity of the RNA. The amount of DNA that hybridizes to rRNA can either be calculated directly from the saturation plateau (Fig. 9.8) or, more accurately, from a double reciprocal plot of the data (Scott & Ingle, 1973). In most plants between 0.05 and 1% of the DNA hybridizes to rRNA. This represents from a few hundred to several thousand genes per diploid nucleus, depending on the species (Table 9.5). The 258 and 18S rRNA molecules hybridize to separate sites on the DNA in accordance with the ratio of their molecular weights (1.3:0.7; approximately 2:1 by mass but 1:1 molar ratio).
Figure 9.8
Saturation hybridization of nuclear DNA from Phaseolus
aureus with radioactive 25S rRNA. 25s rRNA (molecular weight
1.3 × 106 daltons) labelled with [3 H]-uridine at a specific activity
of 1.4 × 104 c.p.m. µg–1 was hybridized to nuclear DNA (2 µg
per filter) for increasing periods of time in 6 × S.S.C. at 70°C.
(From Grierson, 1975).
| |||||||||||||||||||||||||||||||||||||||||||||||||||
This indicates that there are equal numbers of genes for each type of rRNA. In cases where they do not hybridize in equimolar amounts this may be due to sequence homology between the two RNAs, or alternatively may arise by contamination of the RNA samples with other types of RNA.
Multiple genes for rRNA are a physiological necessity in order to provide sufficient sites of synthesis for rRNA molecules to meet the high metabolic demand for ribosomes. Plants often contain thousands of these genes although the number varies between varieties of the same species and between different species. Most of the available evidence suggests that all the copies are identical and this provides a clear example of repetitive DNA with a known function.
Their localization within the nucleolus has been confirmed by in situ hybridization of rRNA to cytological preparations (Brady & Clutter, 1972).
In many eukaryotes ribosomal DNA occupies a separate position in caesium chloride gradients relative to main band DNA. However, since they represent 1%, or less, of the genome these genes are not easily detected by studying the distribution of the DNA. Nevertheless, they can readily be pinpointed by hybridizing rRNA with separate fractions of DNA from the gradient. The fractionation of main band DNA from the rRNA genes of Phaseolus aureus is shown in Fig. 9.9. In general, ribosomal DNA from plants has a higher density
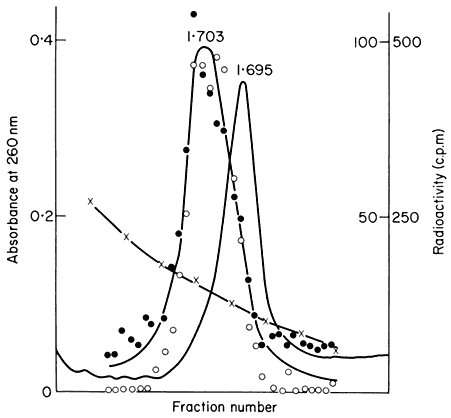
Figuer 9.9.
Separation and detection of the DNA sequences coding for
ribosomal RNA. Nuclear DNA from Phaseolus aureus was
fractionated in caesium chloride (see Figs. 9.4 and 9.5) and
the fractions from the gradient were divided into two and
hybridized to either 25s (cytoplasmic) or 23s (chloroplast)
rRNA labelled with [3 H]-uridine. Smooth, curve, absorbance
of DNA at 260 nm; solid circles, hybridization with 25s rRNA;
open circles, hybridization with 23s rRNA. The gradient of CsCl
is shown and the calculated densities of the rRNA genes and
the main band DNA are indicated. (Unpublished result of D. Grierson.)
than main band DNA. This is consistent with the fact that in higher plants rRNA has a base composition in excess of 50% GC, whereas the bulk of nuclear DNA often has a lower GC content. In this sense rRNA genes represent a type of satellite DNA. However, in different plants the rRNA genes are often present in similar numbers and with a similar density irrespective of the presence or absence of large amounts of satellite DNA. It therefore appears that ribosomal DNA and other satellites may occasionally band together in caesium chloride but this is entirely fortuitous (Ingle et al., 1975).
9.5.2—
Electron Microscopy
The rRNA genes can also be studied by electron microscopy of preparations from isolated nucleoli. This approach provides a clear indication of their organization. Figure 9.10 shows an electron micrograph of the partially
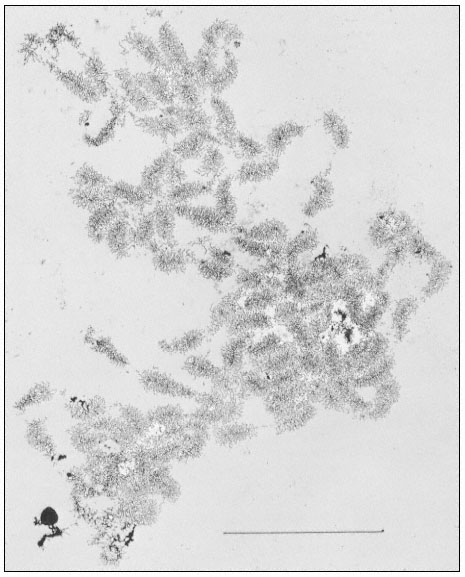
Figure 9.10
Spread contents of a complete nucleolar aggregate subunit from Acetabularia
mediterranea. Note the numerous 'matrix' units. The scale indicates 5 µm.
(From Spring et al., 1974.)
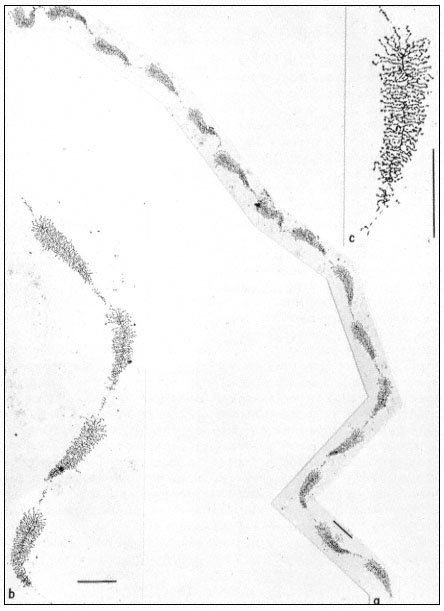
Figure 9.11
(a) Well spread nucleolar material showing the regular pattern of 'matrix'
units and 'spacer' segments. Note the length gradient of lateral fibrils within
each matrix unit. (b) Note occasional groups of small fibrils associated with the
spacer units. (c) At higher magnification, the dense packing of lateral fibrils within
the matrix unit is clearly seen. Scales indicate 1 µm, (From Trendelenburg et al., 1974.)
dispersed contents of a subunit from the nucleolar aggregate of Acetabularia. This shows a regular arrangement of fibril-covered 'matrix' units and fibril-free 'spacer' regions arranged along a central axis. A higher magnification of a 'matrix' unit is shown in Fig. 9.11. The central axis is the double-stranded ribosomal DNA itself. The 'matrix' units represent ribosomal genes which are being transcribed into RNA at a number of points simultaneously. The fibrils of the 'matrix' unit are growing chains of rRNA precursor molecules (see the section on RNA synthesis). RNA polymerase enzymes can be seen clustered on the DNA. About 120 enzymes are engaged in the simultaneous transcription of the DNA in each 'matrix' unit. The marked polarity reflects the fact the growing chains of attached RNA molecules increase in length as the polymerase enzymes responsible for their synthesis proceed along the DNA axis. In addition there is a tendency for the free ends of the RNA chains to form terminal knobs as they become folded, probably in association with proteins.
Multiple rRNA genes therefore alternate in a precise arrangement with non-transcribed regions of the DNA. Hybridization evidence shows that separate 'cistrons' for the 25s and 18s rRNA are present in the DNA, but there is no evidence for the existence of separate transcription units. In contrast, in Acetabularia each transcribed region of double-stranded DNA is approximately 2 µm long. This indicates that it codes for an RNA molecule with a molecular weight of approximately 2 × 106 daltons. This is the combined mass of the 25s and 18s rRNA molecules (1.3 + 0.7 × 106 daltons). There is clear evidence (discussed in the section on RNA synthesis) that ribosomal RNA is first transcribed as a polycistronic precursor molecule. In many plants this precursor has a molecular weight of approximately 2.4 × 106 daltons, and contains one sequence of 25s RNA plus one sequence of 18s RNA together with some extra sequences which are subsequently removed. In order to reconcile such a mode of synthesis with studies on the organization of the rRNA genes it is necessary to propose that each 'matrix' unit consists of an 18s and 25s sequence and some transcribed 'spacer' DNA. Fortunately, in cases where the organization of ribosomal DNA has been studied in detail, for example in Xenopus, evidence for precisely this type of arrangement has been obtained (Birnstiel et al., 1968). Furthermore, recent studies with the electron microscope have provided evidence for a common organization of the rRNA genes in all animals examined (Schibler et al., 1975). A model for the repeating unit, which probably applies also to plants, for it is compatible with all the available evidence, is shown in Fig. 9.12.
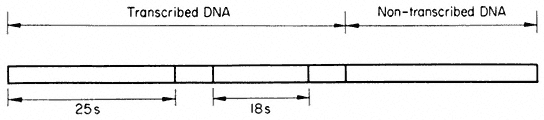
Figure 9.12
Suggested structure of the repeat unit of ribosomal DNA in plants.
(Modified from Schibler et al., 1975.)
By carrying out partial denaturation studies it has proved possible to map certain areas rich in GC or AT base sequences. In addition, it is known that both the non-transcribed 'spacer' regions and the transcribed spacer can vary significantly in length and composition between species. Possible functions for such regions are discussed below. In contrast, however, the regions that code for rRNA are remarkably constant in composition. This probably reflects the importance of their role in providing structural RNA for ribosome formation.
As shown in Fig. 9.9, chloroplast rRNA also hybridizes to nuclear DNA (Ingle et al., 1970; Tewari & Wildman, 1968). This is a surprising result in view of the fact that chloroplasts contain their own DNA and synthesize their own rRNA (see chapter 11). The location of these genes within the nucleus is not known, and at present there is no evidence that they are ever transcribed in the nucleus.
9.6—
Organization and Function of the Genome
9.6.1—
Repetitive DNA
Reassociation studies indicate that between 40% and 90% of the genome of higher plants is composed of repetitive DNA. This fraction is not homogeneous but consists of a number of types of sequence with different lengths and repetition frequencies. In hexaploid wheat, for example, 70%–80% of the genome is made up of families of sequences present in from 100 to 100,000 copies (Smith & Flavell, 1975). The question of the composition, organization and function of these sequences is still under investigation but at least three different classes can be distinguished. These are (a) clustered sequences of repetitive DNA which are transcribed into RNA, (b) clustered sequences which are apparently not transcribed, and (c) reverse-repeats distributed at intervals throughout a substantial part of the genome.
9.6.1.1—
Transcription of Repeated Sequences
DNA-RNA hybridization has shown that a small part of the repeated sequences represents genes for rRNA. These consist of regularly arranged alternating regions of transcribed and non-transcribed DNA. The transcribed sequences can be subdivided into those where the products are stable, the 18s and 25s rRNA; and those that have metabolically unstable products, the transcribed spacer DNA in the rRNA precursor. The role of the transcribed and nontranscribed spacer sequences has not yet been unequivocally established although obvious possibilities are that they may be involved in genetic organization or regulation of transcription. Different groups of rRNA genes may sometimes be present on a number of chromosomes but as far as is known they are all practically identical.
Other families of repeated genes which have been shown to be transcribed include those for 5s RNA, tRNA and, at least in animals, the genes for histone proteins. What proportion of the genome do these account for? The actual value is higher than at first appears because hybridization only detects one strand of the DNA and normally takes no account of either the transcribed or non-transcribed spacer. This means, for example, that a 0.5% hybridization level for rRNA probably accounts for about 2.5% of the total DNA. Similar calculations for the 5s, tRNA and histone genes show that the total number of known sequences of this type only account for something like 5% of the genome.
9.6.1.2—
Palindrome Sequences
A small fraction of rapidly-renaturing DNA is known to be interspersed throughout a large part of the genome. The evidence for this comes from the 'zero-time binding' of DNA to hydroxyapatite. When DNA is denatured and immediately passed through hydroxyapatite a fraction of the DNA binds, suggesting that it is double-stranded. This happens even at very low Cot values where reassociation probably does not occur. The most convincing explanation of this behaviour is that it is due not to strand reassociation but to intra-strand 'snap-back'. This would occur if double-stranded DNA contained reverse-repeat sequences of the following type:
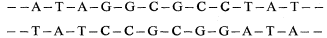
With this arrangement, termed a 'palindrome' sequence, the potential complementarity between adjoining regions of the same single-stranded DNA molecule results in rapid hairpin formation. In consequence any single-stranded DNA molecule will bind to hydroxyapatite providing it contains a double-stranded hairpin region. This provides a means of deducing the organization of palindrome sequences in the genome. The amount of DNA that binds to hydroxyapatite at zero time is markedly dependent upon the fragment size of the DNA used. This suggests that the palindrome sequences are interspersed with other regions of DNA. It has been calculated that in wheat the average distance between these palindrome sequences is 8,500 nucleotide pairs. Between 4% and 10% of the DNA is palindromic and such sequences are spread throughout at least 40% of the genome (Smith & Flavell, 1975).
There are several possible functions for such reverse-repeats. Any RNA molecule transcribed from DNA interspersed with sequences of this type would be expected to have double-stranded hairpin regions. These are known to occur in RNA but the proportion of palindromes in DNA is probably too high to be accounted for solely on this basis. One obvious property of palindromes that may have functional significance is that of symmetry. This may be important
for the recognition of particular regions of double-stranded DNA by enzymes or regulatory elements.
9.6.1.3—
Other Types of Repetitive DNA
Satellite DNAs also consist of repeated sequences. It is clear from Table 9.3 that satellites vary substantially in buoyant density, and therefore base composition, in different plants. This variation is also reflected in sequence content and repetition frequency. Although apparently homogeneous in caesium chloride the melon satellite has been shown to consist of at least two components, judged by melting experiments, caesium sulphate centrifugation and renaturation behaviour. Preliminary estimates are that the repeat lengths of the two sequences are respectively several hundred and a million or more nucleotide pairs (Bendich & Anderson, 1974; Sinclair et al., 1975). It is of interest to ask to what extent the copies within one family of repeated sequences diverge in sequence because this knowledge may provide clues about the origin and function of repetitive DNA. Indirect information on this point comes from melting experiments carried out with native and renatured satellite DNA. When similar, but not identical, sequences renature the possibility exists for the formation of duplexes with a certain proportion of mis-matched bases. For every 1% mismatching in renatured DNA the Tm is decreased by 1.0 ± 0 3°C below that of native DNA (Britten et al., 1974). Judged by this criterion the sequences within the two types of melon satellite DNA are remarkably similar. Many animal satellite DNAs appear to have evolved by duplication of a rather short sequence of DNA followed by divergence from the basic sequence. They are believed not to be transcribed. In contrast, the melon satellite DNAs appear to show very little sequence divergence and furthermore one component at least has a suprisingly large potential coding capacity. However, this may prove to be exceptional and further experimental work is necessary before it can be established whether or not plant satellites are more complex than those found in animals and whether they are ever transcribed into RNA.
The division of repetitive DNA into satellite and non-satellite components is probably an arbitrary distinction. All plants contain repeated sequence DNA irrespective of whether or not they possess satellites. This emphasises the considerable variation in properties of repeated DNA sequences. However, very little is known about their function. The fact that they sometimes form satellites suggests that they are clustered to some extent, otherwise they would remain attached to, and therefore band in association with, main-band DNA in caesium chloride. In the mouse, hybridization to cytological preparations has shown that satellite sequences are concentrated around the centromeres of many of the chromosomes, but this is not always true of satellite DNA.
Where satellites are present the amounts may vary in different cell types of the same plant (Ingle & Timmis, 1975). Furthermore there is some evidence that at least part of the repeated DNA can be dispensed with altogether. For
example, Beridze (1972) has investigated six species of Phaseolus and shown that three contain substantial amounts of satellite DNA whereas three others possess little if any satellite. Clearly, in these cases, the satellite is not essential and its presence or absence appears to have little effect on the plant phenotype. It is quite possible that at least some of the repetitive sequences in DNA are manifestations of evolutionary 'doodling'.
9.6.2—
Unique DNA Sequences
One might conclude that the non-repetitive portion of the genome represents that fraction of the DNA that codes for proteins. However, taking into account the repetitive DNA fraction, the amount of unique sequence DNA in different species is found to vary at least ten-fold (Table 9.4). One way to estimate the proportion of DNA transcribed is to carry out hybridization experiments between mRNA and unique-sequence DNA. Due to the very high Cot values required the rate and extent of hybridization using the conventional filtered technique are unsatisfactory. An alterative approach is to carry out DNA renaturation at high concentrations, using only a trace of highly radioactive RNA for hybridization (Bishop et al., 1974). However, another technical difficulty is encountered in obtaining RNA preparations sufficiently enriched with mRNA and of sufficiently high specific radioactivity for this purpose and no estimates are yet available concerning the extent of transcription of the unique fraction of the plant genome.
9.7—
RNA Synthesis
Most types of RNA are single-stranded molecules transcribed by RNA polymerase enzymes from one of the two strands of a DNA template. RNA molecules are, therefore, the direct products of genes. The following section deals with the transcription of nuclear DNA and the processing and transport of RNA to the cytoplasm. The transcription of chloroplast and mitochondrial DNA and the special case of the synthesis of RNA on a RNA template by plant viruses are excluded.
9.7.1—
General Properties of RNA
Different types of RNA are characterized by their subcellular distribution and properties such as molecular weight and nucleotide composition. Approximately 70% of cellular RNA is contained in the ribosomes where it plays a structural role. Each 80s cytoplasmic ribosome contains three different types of ribosomal RNA (rRNA). These molecules are most commonly referred to by 's values' which are related to their rates of sedimentation during ultracentrifugation.
The larger ribosomal subunit contains two rRNA molecules of 25s and 5s and the smaller subunit contains a single rRNA of 18s. The approximate molecular weights are 1.3 × 106 daltons for 25s rRNA (about 3,900 nucleotides), 0.7 × 106 daltons for 18s rRNA (about 2,100 nucleotides) and 4 × 104 daltons for 5s RNA (about 120 nucleotides). The nucleotide compositions of a range of rRNAs are shown in Table 9.6. Transfer RNA (tRNA or 4s RNA) accounts for a
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
further 25% of the total. In contrast to rRNA this fraction is heterogeneous. It consists of a range of similar but not identical molecules approximately 80 nucleotides long, each capable of being charged with a specific amino acid. By interacting with messenger-RNA appropriately charged tRNAs are responsible for aligning amino acids in the correct order during protein synthesis. In addition to the four major bases tRNA contains a very high proportion of modified or unusual bases. The most interesting type of RNA, mRNA, is present in small amounts and constitutes less than 5% of the total. It is this fraction that directs the synthesis of specific proteins by the ribosomes. The length and nucleotide
sequence of mRNAs are related to the properties of the proteins for which they code. As most plant cells synthesize a variety of different proteins the mRNA fraction generally consists of a range of molecules of different size and nucleotide composition. The fractionation, by polyacrylamide gel electrophoresis, of total RNA from suspension cultures of sycamore cells is shown in Fig. 9.13a. The rRNA and tRNA are clearly distinguished but the small amount of mRNA present is not detected. In Fig. 9.13b purified radioactive mRNA from polyribosomes of sycamore cells is compared with non-radioactive rRNA. The distance moved by each RNA through the gel is, with a few exceptions, inversely related to the log of the molecular weight (Loening, 1968, 1969).
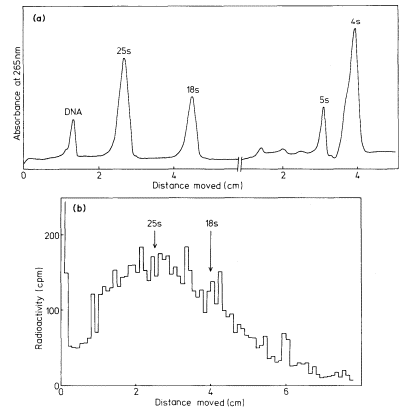
Figure 9.13
The fractionation of sycamore (Acer pseudoplatanus ) RNA by gel
electrophoresis. (a) Composite of two gel profiles. On the left the
fractionation of DNA and 25s and 18s rRNA in a 2.4% acrylamide
gel. On the right the separation of 5s from tRNA in a 7.5% gel. (b)
Gel electrophoresis of poly (A)-containing mRNA purified from
polyribosomes of sycamore cells labelled with [3 H]-uridine for
30 minutes. The positions of unlabelled 25s and 18S rRNA used
as molecular weight markers in the same gel are indicated.
(Unpublished results of D. Grierson.)
9.7.2—
Ribosomal RNA Synthesis
9.7.2.1—
The Ribosomal RNA Precursor
The DNA that codes for rRNA is known to be associated with nucleoli and the transcription of nucleolar DNA has been studied with the electron microscope (Fig. 9. 11). More detailed information has been obtained by the labelling of intact seedlings, plant segments, or cultured cells, with [32 P]-phosphate or [3 H]-uridine for various times followed by the extraction and characterization of the rapidly-labelled RNA. The mechanism of rRNA synthesis is fairly well understood and a generalized scheme outlining the main features of the process is given in Fig. 9.14. The first stable gene product which can be detected is an rRNA precursor molecule. The molecular weight of this RNA varies in different plants but is generally within the range 2.2–2.6 × 106 daltons. The rRNA precursor from mung bean roots, detected by polyacrylamide gel electrophoresis,
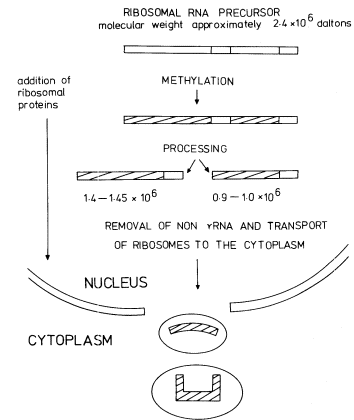
Figure 9.14
The synthesis and processing of the precursor to rRNA.
(Modified from Grierson et al., 1975.)
is shown in Fig. 9.15. The precursor undergoes considerable post-transcriptional modification in the nucleolus before it is converted into 'mature' rRNA and enters the cytoplasm. Shortly after synthesis the molecule becomes methylated at selected sites. This can be demonstrated by incubating plants in methyl-labelled methionine for a short time. The methyl groups become incorporated into the rRNA precursor via s-adenosyl methionine (Cox & Thurnock, 1973). Ribosomal-RNA is known to contain 2'-methyl ribose at certain positions. Only a small number of the residues are modified in this way and it is probable that methylation is completed during the rRNA precursor stage.
Figure 9.15
Gel electrophoresis of total RNA from roots of P. aureus labelled for
1.5 h with [32 P]-phosphate. The molecular weights of the rRNA precursor and
processing intermediates are shown in millions. (Unpublished result of D. Grierson.)
The precursor molecules correspond to the lateral fibrils attached to the matrix units of active nucleolar DNA (Fig. 9.11). Each molecule contains one sequence of 25s rRNA, one sequence of 18s (total molecular weight = 2.0 × 106 daltons) together with some non-rRNA. The evidence for this is as follows. Firstly, although it is rapidly labelled, the precursor does not accumulate in large amounts. This suggests, therefore, that it must either be rapidly degraded or converted to some other molecular form (Rogers et al., 1970; Leaver & Key, 1970; Grierson et al., 1970; Cox & Turnock, 1973; Grierson & Loening, 1974). This latter explanation would account for the fact that radioactivity appears in mature rRNA slightly later than in the precursor. Secondly, the nucleotide composition of the rRNA-precursor is similar to that of rRNA (Rogers et al., 1970; Leaver & Key, 1970; Grierson & Loening, 1974). Thirdly, competition hybridization experiments have shown that both unlabelled 25s and 18s rRNA compete with radioactive precursor-RNA for sites in the DNA, suggesting that
they share common sequences (Fig. 9.16). Finally, partial digestion of radioactive 25S, 18s and precursor RNA by T1 ribonuclease, followed by two-dimensional fractionation of the fragments shows that the 'fingerprint' pattern of the precursor is similar to that expected of a mixture of 25s and 18s rRNA (Sen et al., 1975).
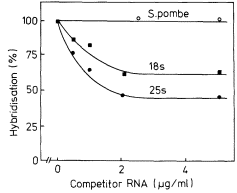
Figure 9.16
Competition hybridization between unlabelled
rRNA and radioactive rRNA precursor for
similar sites in P. aureus DNA.
(From Grierson & Loening, 1974.)
In some plants more than one precursor is detected (Leaver & Key, 1970; Cox & Turnock, 1973). The two precursors from cultured sycamore cells are shown in Fig. 9.17. In some instances the second precursor may originate in chloroplasts or mitochondria (Grierson & Loening, 1974; Kuriyama & Luck, 1973), but in carrot and sycamore both precursors are thought to be in the nucleus (Leaver & Key, 1970; Cox & Turnock, 1974). It is not clear in these cases whether the two RNAs contain similar sequences or whether they are transcribed from different genes. It is possible that the larger RNA is converted to the smaller one, but careful analysis of the rate of labelling of both molecules does not support this suggestion (Cox & Turnock, 1973; Leaver & Key, 1970).
9.7.2.2—
Processing of the Precursor
In addition to becoming methylated, and while still in the nucleolus, the precursor becomes associated with protein molecules and is processed by nucleolytic enzymes which remove the excess non-rRNA in stages to produce mature rRNA in ribosomal subunits ready for transport to the cytoplasm. Some RNA processing intermediates are present in the sample shown in Fig. 9.15. The scheme for processing outlined in Fig. 9.14 is consistent with some, but not all, the studies on rRNA synthesis in plants. For example, no immediate precursor to 25s rRNA is detectable in rye embryos (Sen et al., 1975). Furthermore the 1.0 × 106 daltons RNA is not always detected wheras certain additional RNAs
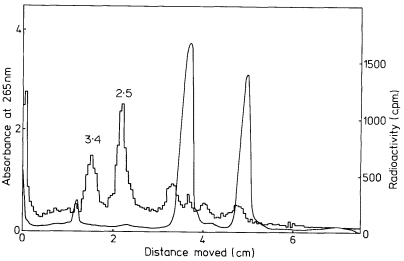
Figure 9.17
Pulse-labelled RNA from sycamore cells showing two rRNA precursor
molecules. The cells were labelled for 40 minutes with [3 H]-uridine and
total nucleic acid fractionated by gel electrophoresis. The DNA was not
removed but an identical distribution of radioactivity is observed when
this is carried out. The approximate molecular weights of the precursor
RNAs are indicated in millions. (Unpublished result of D. Grierson.)
are sometimes observed. For example, after cycloheximide treatment of cultured parsley cells, which slows down rRNA processing, previously undetected RNAs with molecular weights of 2.0 and 0.9 × 106 daltons were found (Gebauer et al., 1975). This suggests that if certain processing events occur very rapidly they may normally go undetected. It is very probable that the complete sequence of processing steps will only be worked out when it becomes possible to study accurately RNA metabolism in isolated nucleoli. Apparent differences between closely related species may be explained by assuming that certain critical stages of processing occur at different rates. In addition, it should be realised that the processing enzymes are functioning at different, possibly distant, sites within the same molecule. There seems no compelling reason to expect these modifications always to occur in the same chronological order and, if they do not, variations in the pattern of processing intermediates might be expected under altered conditions or in different species.
The function of the excess RNA in the precursor is not known. It is too variable in size in different organisms to be a mRNA for the ribosomal proteins (Loening, 1970) and in any case it appears to be degraded very rapidly after synthesis. It may play some role in the regulation of transcription but more probably it is necessary for ensuring that rRNA adopts the correct secondary and tertiary structure required for methylation and for the addition of proteins during ribosome assembly.
It is pertinent to ask whether the multi-stage mechanism that operates for rRNA production is unique or whether it represents a general principle common to the synthesis of all types of RNA. Precursors to tRNA and 5s RNA have been identified and characterized in bacteria and animals (Hecht et al., 1968; Bernhardt & Darnell, 1969) and precursors of mRNA have also been detected, although there is uncertainty about the actual details of processing (Brawerman, 1974). Although RNA metabolism has not been so intensively studied in plants the indications are that precursors to 5s RNA, tRNA and mRNA do occur. When considering the question of the regulation of RNA metabolism, therefore, it is necessary to take account of post-transcriptional modifications as possible control points.
9.7.3—
Messenger-RNA Synthesis and Metabolism
The question of mRNA synthesis has proved extremely difficult to investigate due largely to problems of identification and purification of mRNA. In the past, rather imprecise criteria for mRNA identification, such as size heterogeneity, similarity in nucleotide composition to DNA, rapidity of labelling and association with polyribosomes, were adopted. More recently, two additional approaches have become available. Firstly, the development of an in vitro protein synthesizing system has made it possible to test the capacity of an RNA fraction to direct the synthesis of specific proteins. Secondly, the discovery that many mRNAs contain sequences of poly (adenylic) acid (poly (A)) has provided a simple approach to the purification of mRNA.
9.7.3.1—
Poly (A)-Containing RNA
It has been known for some considerable time that a small fraction of total plant RNA has a very high AMP content. This fraction, often referred to as DNA-like RNA or D-RNA, is considered to consist, at least in part, of mRNA and is partially purified by chromatography on columns of methylated albumin adsorbed to kieselguhr (Ingle et al., 1965; Lin et al., 1966; Jackson & Ingle, 1973). More recent work with animal mRNA has shown that the high AMP content is due to the presence of regions of poly (A) in the mRNA molecules. These sequences of poly (A) are generally 50 to 250 nucleotides in length. In animals they are attached to the 3' end of the mRNA molecules after transcription (Brawerman, 1974) although there is some evidence that shorter regions of poly (A) may also be introduced into RNA as part of the transcription process (Jacobson et al., 1974). Poly (A)-containing RNA has been identified in a variety of animals (Brawerman, 1974; Matthews, 1973) and plants (Sagher et al., 1974; Verma et al., 1974; Higgins et al., 1973) although it has not been found in higher plant chloroplasts (Wheeler & Hartley, 1975). It is apparently present in mitochondria from some organisms (Perlman et al., 1973; Avadhani et al.,
1973; Ojala & Attardi, 1974) but not others (Groot et al., 1974). This may reflect true differences between organisms or alternatively may be a question of the experimental technique. For example, contrary to earlier reports that poly (A) is absent from mRNA from bacteria, it has recently been shown to be present at a low level in E. coli. (Nakazato et al., 1975).
The function of the poly (A) sequences is not known but their presence can be used to purify mRNA. There are a number of methods for doing this. Poly (A) will bind to nitrocellulose or cellulose at high salt concentrations. RNA without poly (A) can be washed off and the poly (A)-containing RNA is then eluted at low ionic strength. However, cellulose and nitrocellulose do not necessarily retain all the poly (A)-containing RNA. A more specific type of binding is achieved using columns of oligo (deoxythymidylic) acid attached to cellulose or poly (uridylic) acid attached to sepharose. Both these polymers rapidly form stable nucleic acid hybrids with the poly (A) sequence attached to the mRNA. Other types of RNA pass straight through the column, although it is sometimes necessary to wash through extensively with a buffer. The poly (A)-containing RNA is only released after lowering the salt concentration or raising the temperature to cause melting of the hybrid between poly (A) and either oligo (dT) or poly (U).
There is considerable evidence that poly (A)-containing RNA from plants is mRNA. It is associated with polyribosomes (Higgins et al., 1973; Grierson et al., 1976) and its synthesis is inhibited by the antibiotic a -amanitin which prevents transcription of non-rRNA in the nucleus (Higgins et al., 1973). When purified from cells synthesizing a large number of proteins of different sizes poly (A)-containing RNA is polydisperse, but when extracted from soybean root nodules, which synthesize substantial amounts of leghaemoglobin, two peaks of RNA are detected, each presumably consisting of similar or identical RNA molecules. These RNA fractions stimulate protein synthesis by a cell-free system from wheat germ. Furthermore, leghaemoglobin has been positively identified among the products of in vitro synthesis (Verma et al., 1974). Supporting evidence that poly (A)-containing RNA stimulates protein synthesis comes from a large number of studies and the small number of individual proteins identified as in vitro products is growing steadily. However, it should be stressed that not all mRNA molecules contain poly (A). In animals, for example, about 70% of mRNA is estimated to contain poly (A) (Milcarek et al., 1974) whereas only about 40% of polydisperse RNA from polyribosomes of cultured sycamore cells can be purified by oligo (dT)-cellulose (Covey & Grierson, 1976). The functional significance of the two mRNA populations is not clear.
9.7.3.2—
Postranscriptional Modification of mRNA
Poly (A)-containing RNA is synthesized in the nuclei of higher plant cells. Total rapidly-labelled RNA from nuclei consists of the rRNA precursors and pro-
cessing intermediates together with polydisperse RNA (Fig. 9.18). Approximately 15% of this RNA is retained by oligo (dT)-cellulose and by this criterion is judged to contain poly (A) sequences (Grierson & Covey, 1976). It is polydisperse in size and has a high AMP content (Table 9.6). There is little direct evidence concerning the mechanism of poly (A) addition to nuclear polydisperse RNA in plants. It is assumed to be added to the 3' end of the molecules by the enzyme ATP-poly-nucleotidylexotransferase after transcription by RNA polymerase (Maale et al., 1975). The poly (A) sequences in higher plant RNA have been estimated to range from 50–250 nucleotides in length (Sagher et al., 1974; Covey & Grierson, 1976). However, plant DNA appears to contain short sequences of AT base pairs which could give rise to short poly (A) regions by transcription (Grierson, 1975) and there is evidence that similar regions are transcribed in slime moulds (Jacobsen et al., 1974).
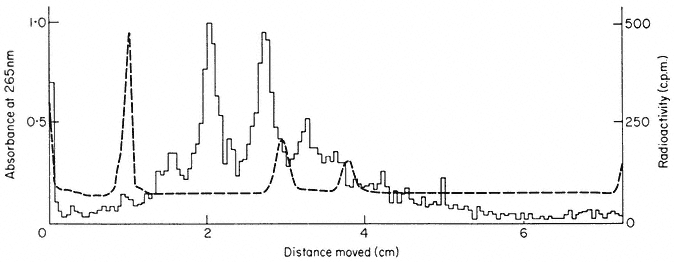
Figure 9.18
Gel electrophoresis of total nucleic acid from nuclei of sycamore cells.
The cells were labelled for 1 h with [3 H]-uridine, a nuclear fraction was
prepared and total nucleic acid extracted and fractionated by gel
electrophoresis. The radioactivity is shown as a histogram.
Note that the nuclear fraction contains very
little rRNA by comparison with Fig. 9.17.
(Grierson & Covey, 1976.)
Comparison of the nuclear and cytoplasmic poly (A)-containing RNA by gel electrophoresis suggests that nuclear RNA is reduced in size before entering the cytoplasm (Fig. 9.19). This suggests that there may be a processing mechanism analogous to that which operates for rRNA synthesis. In slime moulds the nuclear precursor appears to be approximately 20% larger than cytoplasmic mRNA. (Firtel & Lodish, 1973). In higher plants the average molecular weight of the nuclear poly (A)-containing RNA is 1 × 106 daltons (Grierson & Covey, 1976). This is in contrast to the giant precursors to mRNA earlier claimed to occur in animal cells. Very little is known about the processing mechanism, but Firtel and Lodish (1973) have shown that the nuclear precursor contains a sequence of about 300 nucleotides near the 5' end of the molecule which is lost before the RNA enters the cytoplasm. This is of considerable interest
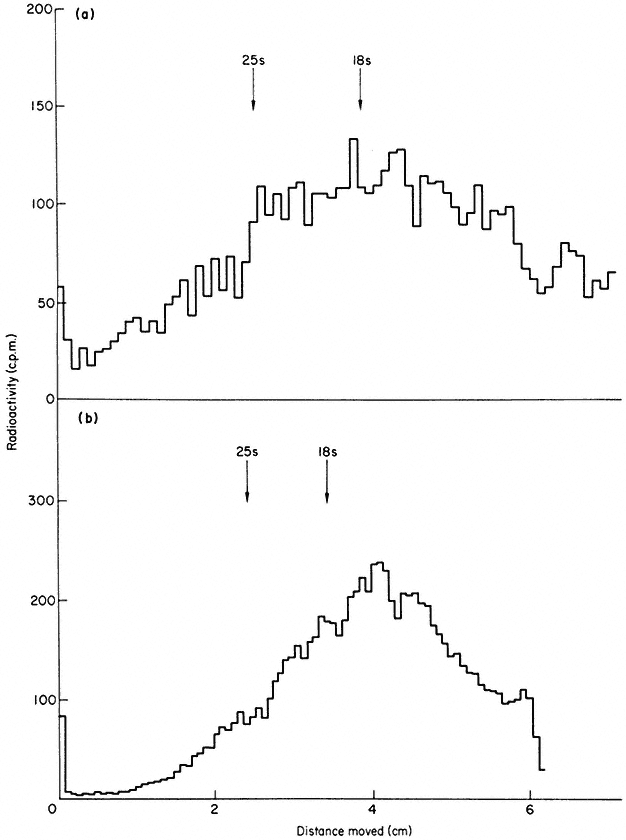
Figure 9.19
Gel electrophoresis of poly (A)-containing RNA from nucleus and cytoplasm.
Pulse labelled sycamore cells were separated into (a) nuclear, and (b) cytoplasmic
fractions and poly (A)-containing RNA purified by oligo (dT)-cellulose chromatograph.
The two RNA preparations were denatured in 8 B urea at 60°C to disrupt aggregates
and fractionated by gel electrophoresis in the presence of marker rRNA. The cytoplasmic
RNA is distinctly smaller, on average, than the nuclear RNA. (Grierson & Covey, 1976.)
because this region appears to be transcribed from repetitive DNA. It therefore seems that some of the repeated DNA sequences interspersed between nonrepetitive DNA are transcribed and may play a regulatory role in processing or transport of mRNA.
9.8—
Nucleic Acid-Protein Interactions
9.8.1—
Histones
Chromosomal DNA is invariably associated with several different types of proteins (Table 9.7). These include histones which occur in most eukaryotes.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
There are five main histone proteins designated F1 , F2b , F2a1 , F2a2 and F3 , with molecular weights from 11,000 to 22,000 daltons. They are strongly basic and generally have a high content of arginine and lysine. The basic amino acids are often clustered towards either the N or C terminus of the proteins (Macgillivray & Rickwcod, 1974) suggesting that these regions interact directly with the DNA. Stedman and Stedman (1950) proposed that histones are responsible in some way for the regulation of gene expression but it now seems that they lack the specificity for recognition of different gene sequences and it is more likely that they fulfil a more general role in the organization of chromosomes. Comparison of the amino acid sequences of histones from different species suggests that they have been highly conserved during evolution. For example, the sequences of F2a1 from calf, pig and rat are identical and differ only in two substitutions from F2a1 from pea (Delange et al., 1966). This suggests these proteins evolved to fulfil an essential role with strict requirements
for a precise sequence, although other histones, notably F2b and F2a2 show greater variability. Histones frequently undergo modifications such as methylation, acetylation or phosphorylation. These are normally reversible and would be expected to alter the association of histones with DNA. In general these modifications occur at about the time of DNA synthesis although they may also herald an increase in RNA synthesis. During the cell cycle the synthesis of histones is closely linked to that of DNA and phosphorylation of F1 has been shown to precede cell division (Bradbury et al., 1974). Basic non-histone proteins and acidic proteins also occur in association with DNA. These form a more heterogeneous group than the histones and include components of different molecular weight, charge and tissue distribution. They are less stable metabolically than histones and do not behave as a single class of proteins. Several lines of evidence suggest that this group may include regulatory elements (Macgillivray & Rickwood, 1974).
9.8.2—
Subunit Structure of Chromatin and Chromosome Organization
The double helical structure of DNA is widely known and it is equally widely recognized that a further degree of organization is responsible for the folding of 1 m or more of DNA into each set of chromosomes. Early observations on the structure of chromosomes indicated that DNA is organized in fibres of the order of 10 nm thick and it was suggested that the double helix, in association with proteins, is supercoiled. In addition, X-ray studies suggested that there is a repeating structure in chromatin at intervals of approximately 10 nm and more recently a linear arrangement of beads of the order of 7 nm diameter has been observed in chromatin with the aid of the electron microscope (Olins & Olins, 1974). Further evidence for a regular arrangement of DNA in chromatin comes from the observation that following nuclease digestion reproducible fragments of DNA approximately 205 base pairs long are produced (Noll, 1975). Kornberg (1974) has suggested a repeating subunit structure of approximately 200 DNA base pairs associated with 2 molecules of each of the four major histones (F2b , F2a1 , F2a2 and F3 ) and one molecule of F1 . In this model the DNA double helix is envisaged to be folded to approximately one seventh its length and Crick and Klug (1975) have suggested that this could occur by 'kinking' of the DNA helix at intervals of perhaps 20 base pairs. Bradbury and his colleagues (Baldwin et al., 1975) have proposed that repeating units of DNA approximately 200 base pairs long are coiled on the outside of aggregates of the four major histones which associate with each other by apolar interactions, leaving their basic regions free to associate with the DNA. Histone F1 , which is believed not to be directly involved in the subunit, may perform a cross-linking function. Only about 87% of the DNA in chromatin is protected against deoxyribonuclease digestion (Noll, 1975) and it is possible that not all the DNA participates in formation of this type of subunit.
Further insight into the functional organization in chromatin comes from studies on the giant chromosomes from certain insects. These chromosomes consist of a series of densely coiled bands connected by less dense regions. These are much larger than the repeating unit of 200 DNA base pairs. Each band plus interband region is considered to correspond to a gene because there is good numerical agreement between their number and the number of complementation groups determined by breeding experiments. However, calculations show that only a fraction of the DNA in each band plus interband region is necessary in order to code for a protein (Beerman, 1972) and the question of the function and organization of the DNA remains unresolved. Regions of the DNA being actively transcribed appear as chromosome puffs composed of DNA, RNA and protein. Crick (1971) has suggested that the interband regions contain DNA that is transcribed and that each band consists of regulatory DNA sequences. In contrast, Paul (1972) has proposed a different type of arrangement.
Different regions of chromosomes often contain chromatin condensed to different extents (Macgillivray & Rickwood, 1974). Euchromatin appears to be loosely packed. It is known to contain both unique and repeated sequences and is actively transcribed. Heterochromatin is more tightly packed and consists of two types. Constitutive heterochromatin contains sequences that are never transcribed and includes repeated sequences. In some species satellite DNAs are known to be clustered in heterochromatic regions near the centromeres. Finally, facultative heterochromatin constitutes those regions of the chromosome that sometimes appear as euchromatin.
9.8.3—
Regulation of Transcription
Thus there is evidence for the coiling of DNA and formation of regular DNA-protein complexes which may be further folded into larger units. Different regions of the chromosome are folded to different extents and this presumably affects the availability of DNA for transcription. Comparison of the extent of transcription of pure DNA and isolated chromatin by added bacterial RNA polymerase suggests that in animals only about 10% of the genome is available for RNA synthesis. Hybridization experiments suggest that the RNA synthesized from isolated animal chromatin is tissue specific and resembles in sequence the type of RNA synthesized in vivo (Macgillivray & Rickwood, 1974). It is probable that the non-histone proteins are more important in determining which segments of the genome are repressed but further characterization of the properties and mechanism of action of these proteins is necessary before their function becomes clear (see Mcgillivray & Rickwood, 1974). It has also been suggested that chromosomal RNA may be important in regulating gene expression, possibly in conjunction with repeated sequences in DNA (Georgiev, 1969; Ryskov et al., 1971; Britten & Davidson, 1969). There is undoubtedly RNA associated with chromatin, including growing chains of polydisperse-
RNA and rRNA-precursor, but the existence of an additional type of regulator RNA has been proposed. However, at the present time there is a certain amount of controversy concerning the characterization and properties of chromosomal RNA (Jacobson & Bonner, 1971; Huang, 1967; Heyden & Zachau, 1971; Holmes et al., 1972). As yet there is no evidence that this RNA regulates transcription.
In addition to repression of the genome by interaction with proteins, template selection may also be controlled by RNA polymerase enzymes and by other protein factors that interact with the polymerases to regulate transcription. As far as studies on plant chromatin are concerned in vitro RNA synthesis has been achieved using the endogenous RNA polymerases that remain attached to the isolated chromatin (Guilfoyle & Hanson, 1974). However, there have been few attempts to characterize the RNA products in detail. There are several distinct RNA polymerases with different properties. These enzymes are probably responsible for transcribing different regions of the genome. For example, RNA polymerase I, which is insensitive to the antibiotic a -amanitin, is located within the nucleolus (Brandle & Zetsche, 1973). This enzyme appears to be responsible for transcription of the rRNA genes. The activity of this enzyme and the rate of RNA production are significantly enhanced in tissue treated with auxin (Guilfoyle et al., 1975). Enzymes sensitive to a -amanitin are present in the nucleoplasm and presumably are responsible for transcribing other parts of the genome. These polymerases may also be affected by hormones via the mediation of hormone-binding factors (see chapter 13). In addition, evidence has been obtained for the existence of other protein factors which govern the activity of RNA polymerases by stimulating initiation of RNA synthesis (Mondal et al., 1972a, 1972b).
9.9—
Conclusions
Now that we have a general idea of the organization and properties of DNA it is possible to pose more precise questions about the function of the different parts of the genome. For example it would be interesting to establish what proportion of the DNA is transcribed into RNA in plants with different nuclear DNA contents. There is also the question of the function of the repeated DNA sequences: are they involved in regulation of transcription; to what extent are they transcribed; and what is the function of the repetitive sequences in the nuclear precursors to mRNA? Further characterization of the various RNA polymerase enzymes and the protein factors which influence RNA synthesis will doubtless increase our knowledge of the control of gene expression. However, it does seem that ultimately such studies should be carried out using chromatin as template in order to understand the precise molecular interactions involved. Finally, the possibility of post-transcriptional regulation should not be overlooked. It is important to establish the role of post-transcriptional modification
of RNA molecules in relation to mRNA selection and transport to the cytoplasm.
Further Reading
Biswas B.B., Ganguly A. & Das A. (1975) Eukaryotic RNA polymerases and the factors that control them. In Progress in Nucleic Acid Research and Molecular Biology (ed. W.E. Cohen), Vol. 15, pp. 145–84. Academic Press, New York.
Bryant J.A. (1976) Nuclear DNA. In Molecular Aspects of Gene Expression in Plants (ed. J.A. Bryant), pp. 1–51. Academic Press, London.
Duda C.T. (1976) Plant RNA polymerases. Ann. Rev. Pl. Physiol.27, 119–32.
Grierson D. (1976) RNA structure and metabolism. In Molecular Aspects of Gene Expression in Plants (ed. J.A. Bryant). pp. 53–108. Academic Press, London.
Kornberg A. (1974) DNA Synthesis. W.H. Freeman & Co., San Francisco.
Lewin B. (1974) Gene Expression, Volume 2, Eukaryotic Chromosomes. J. Wiley & Sons Ltd., London.
Nagl W. (1976) Nuclear organisation. Ann. Rev. Pl. Physiol.27, 39–69.
Parish J.H. (1972) Principles and Practice of Experiments with Nucleic Acids. Longman, London.
Rae P.M.M. (1972) The distribution of repetitive DNA sequences in chromosomes. In Advances in Cell Biology and Molecular Biology (ed. E.J. Dupraw). Vol. 2, pp. 109–49. Academic Press, New York.
Stewart P.R. & Letham D.S. (eds.) (1973) The Ribonucleic Acids. Springer-Verlag, Berlin.
Chapter 10—
Protein Synthesis in the Cytoplasm
10.1—
Introduction
Since gene expression occurs almost exclusively via the synthesis of proteins, an understanding of the mechanism of this process is important. Although living organisms make several thousand different proteins with a variety of functions, present evidence suggests that they are all made, by the same basic mechanism, on polysomes. These structures consist of a template RNA molecule, messenger-RNA (mRNA), with several associated ribosomes, and protein synthesis is a complex, energy requiring, multi-enzymic process. A template mechanism of synthesis is dictated by the fact that the protein product has a specificity which depends upon the precise sequence of the constituent amino acid residues in the polypeptide chain. This automatically rules out the normal enzymic method by which most other cell constituents are made, since each enzyme of a set specific for the synthesis of one protein would, in turn, require a further set for its synthesis, ad infinitum. However, since some small polypeptides are synthesized by a non-template mechanism, it is possible that some proteins with highly repetitive sequences of amino acid residues, may be synthesized similarly.
Because all organisms probably synthesize proteins by a similar mechanism, the tendency has been to use an organism which is particularly amenable for basic studies. This organism is the prokaryote, Escherichia coli, and higher plant material has not been much used because of associated technical difficulties. However, recognition of the importance of the deposition of proteins in plants as our major source of protein food, is leading to an increased spate of work in the desire to understand, and possibly manipulate, this important process in the major food plants.
Proteins are synthesized on free and membrane-bound polysomes in the cytosol or in various sub-cellular organelles, e.g. mitochondria, chloroplasts. Only the biochemistry of protein synthesis by cytoplasmic ribosomes will be reviewed in this chapter; organelle protein synthesis is dealt with in chapter 11. Different aspects of the control of protein synthesis are covered in chapters 13 and 15, but it should be re-iterated here that the level of protein in the cell depends on a dynamic balance between synthesis and degradation.
10.2—
Polysomes
10.2.1—
Structure of Ribosomes
Ribosomes are classified as eukaryotic or prokaryotic in type, on the basis of: (a) sensitivity to various antibiotics; (b) functional interchangeability of soluble factors and ribosomes from different sources; and (c) structure and sedimentation characteristics, e.g. 70s (prokaryotic) or 80s (eukaryotic).
Bacterial ribosomes, plant chloroplast and mitochondrial ribosomes are classified as prokaryotic, even though they may not always exhibit all of the above characteristics, e.g. plant mitochondrial ribosomes have sedimentation values of about 80s.
Plant and animal cytoplasmic ribosomes are of the eukaryotic type and have sedimentation values of about 80s. This class of ribosomes are heterogeneous in size however, with most of the differences accounted for by differences in the large subunit, the small subunit having been conserved during evolution (Cammarano et al., 1972a,b & c).
Measurements of the size of plant cytoplasmic ribosomes in the electron microscope vary, but they are approximately 25 × 20 nm. Miller et al., (1966) have described them as being acorn-shaped, and several workers have described a cleft in the small subunit. In the rat liver ribosome model of Nonomura et al., (1971) there is a tunnel between the two subunits, directly under the cleft in the small subunit, which is thought to accommodate the mRNA. Ribosomes require Mg2+ for structural integrity and dissociate at low Mg2+ concentrations into a large and a small subunit with sedimentation values of about 60s and 40s respectively (Ajtkhozhin et al., 1972). There is good evidence that mRNA attaches to the small subunit before the addition of the large subunit completes the ribosome structure, and the different roles of the two subunits are a feature of all the models of ribosome structure and function (see Noll et al., 1973). The 80s ribosome of plants corresponds to a molecular weight of 3.9 × 106 daltons, with molecular weights for the large subunit of 2.4 × 106 daltons and the small subunits, 1.5 × 106 daltons; the small subunit is of the same molecular weight as its mammalian counterpart, the large subunit is smaller. The large subunit of plant ribosomes contains one molecule of 25s RNA, hydrogenbonded to 1 molecule of 5.8s RNA (Payne & Dyer, 1972), and 1 molecule of 5s RNA; the small subunit contains 1 molecule of 18s RNA. The molecular weight of the 25s RNA is 1.3 × 106 daltons, and that of the 18s is 0.7 × 106 daltons (Loening, 1968). The large subunit contains 46% protein, and the small 54%.
The large subunit of pea seedling ribosomes contains 44–45 proteins and the small subunit 32–40 proteins, of which most are basic and of molecular weights between 20 × 103 and 30 × 103 daltons. Some proteins may be represented by more than one copy per ribosome. Proteins extracted from cytoplasmic
and chloroplast ribosomes of the same species, show little similarity in two-dimensional electrophoresis or by immunological comparison and are significantly less similar than are the cytoplasmic ribosomes of different species, e.g. beans and wheat (Gualerzi et al., 1974).
Before a complete understanding of the mechanism of protein synthesis is elucidated, it will be necessary to know the spatial relationships and three-dimensional structures of the proteins and the RNAs of the ribosome, as well as those of the associated molecules which also play a part in protein synthesis. Information on the structure of the proteins and the RNA molecules of the E. coli ribosome, together with reconstitution experiments, is now well advanced (Traub & Nomura, 1968; Nashimoto et al., 1971; Wittmann, 1973; Anderson et al., 1974; Nierhaus & Dohme, 1974).
10.2.2—
Free and Membrane-Bound Polysomes
Ribosomes associate with mRNA to form polysomes, as can be seen in the electron micrograph (Fig. 10.1); the size of the polysome varies according to the length of the mRNA and the number of attached ribosomes. Polysomes are found either free in the cytoplasm or attached to the surface of membranes of the endoplasmic reticulum (ER) and the nucleus. In animal embryonic cells, most of the polysomes are free and engaged in protein synthesis for internal use, whereas in those differentiated animal cells from which large amounts of protein are exported, most of the polysomes are found attached to the ER. Thus, the generalization arose that membrane-bound ribosomes synthesize protein for export and free ribosomes for intracellular use. It soon became clear however, that membrane-bound ribosomes occur in some tissues which do not export protein, and that in cells where all the protein synthesized is for internal use, different classes of protein are synthesized on the two types of ribosomes. Less information is available for plants. In the developing broadbean seed the highly vacuolate, relatively membrane-free cells of the cotyledon are transformed just prior to the onset of storage protein synthesis, into cells whose cytoplasm is vesicular and in which ER with attached ribosomes is very prominent. This new protein synthesis machinery is assembled at a precise time in the course of seed development for the production, in large amounts, of the storage proteins of the seed (Bailey et al., 1970; see Fig. 10.2). Later, during dehydration of the seed, ribosomes become detached and the population of free ribosomes thereby increased. A similar series of events has been recorded for the developing seeds in many other plants.
The way in which ribosomes attach to membranes is not clear; the ribosomes themselves, the membranes and the protein being synthesized, have all been suggested as being involved in the binding. There is evidence from animals that the large subunit is in close contact with the membranes, and Baglioni et al. (1971) have postulated that the large subunit binds directly to the membrane, probably at a specific binding site (Sunshine et al., 1971), and that a protein on
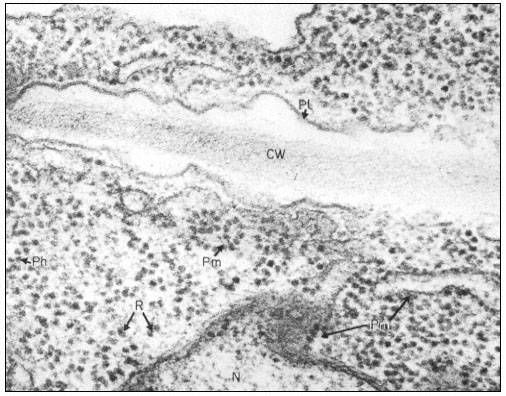
Figure 10.1
Electron micrograph of a thin section of parts of two adjacent cells in a
shoot apex of pea showing ribosomes and polyribosomes; CW = Cell Wall, Pl
= Plasmalemma, N = Nucleus, Pm = Membrane bound polysomes, R = Ribosome
Ph = Polysome helix. By courtesy of A. D. Greenwood, Department of Botany
and Plant Technology, Imperial College of Science and Technology, London.
the membranes is responsible for the attachment (James et al., 1969). Alternatively, it has been suggested that one of the proteins of the large subunit is responsible, whereas other observations suggest that the binding may be dependent on the nascent polypeptide chain. It now seems likelly that, in vivo, membrane-bound ribosomes synthesize a different class of protein to free ribosomes, whereas in vitro, proteins of both classes are synthesized by both types of ribosomes, suggesting the control is not a function of the ribosome itself.
10.2.3—
Isolation and Purification
Ribosomes and polysomes are normally isolated from plants and purified by sedimentation through sucrose cushions as originally described by Wettstein
Figure 10.2
Electron micrographs of developing seeds of Vicia faba.
(a) 25 days after fertilization. Ribosomes free in the cytoplasm. Very little ER present.
et al. (1963). In a typical procedure, the material is homogenized in 0.25 M sucrose containing 200 mM -Tris-HCl, pH 8.5 at 2ºC, 500 mM -KCl and 15 mM -MgC12 , with a Willems Polytron for 3 seconds at a speed setting of 8. The homo-
Figure10.2
(b) 55 days after fertilization, showing most of the polysomes bound to ER and
storage protein being laid down. C = Cytoplasm, P = Protein body, CW = Cell Wall,
RER = Endoplasm reticulum with polysomes attached, N = Nucleus, V = Vacuole.
From Boulter et al. (Qual. Plant. XXIII, 239–50, 1973).
genate is filtered through Miracloth and Triton X-100 is added to a final concentration of 2% (v/v). The filtrate is centrifuged at 104 × g for 10 min at 2°C and the ribosomes and polysomes are recovered from the supernatant by layering over a 3 ml cushion of 0.1 M sucrose in 50 mM -Tris-HCI, pH 8.5, 50 mM -KCl
and 10 mM -MgCl2 , followed by centrifugation for at least 44 x 107 g-min, as described by Leaver & Dyer (1974) (Fig. 10.3). The inclusion of the detergent
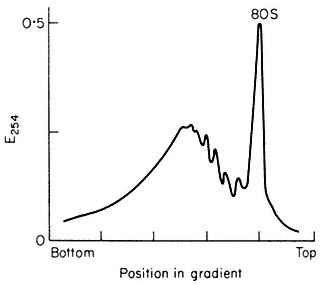
Figure 10.3
Sucrose density profiles of ribosomes and polysomes from Pisum
sativum. Ribosomes recovered after 6 h. centrifugation through
1 M -sucrose cushion. E254 = absorbance at 254 nm
(From Leaver & Dyer Biochem. J. 144, 165–7, 1974.)
Triton X-100 is necessary since plant materials contain a proportion of polysomes which are membrane-bound and the addition of Triton X-100 solubilizes the endoplasmic reticulum, so releasing them. The method gives a preparation of both polysomes and ribosomes and the latter can be removed by centrifugation techniques or, alternatively, the whole preparation can be converted to ribosomes by exposure of the plants to nitrogen gas for at least 1 hour prior to extraction. Since different plants and even different tissues from the same plant contain different amounts of membrane-bound to free polysomes and different free polysomes to free monoribosome ratios, isolation conditions may need to be varied for optimum results with different experimental materials. The situation is further complicated by the fact that plant mitochondrial ribosomes, although not functionally of the 80s type, sediment at 80s (Leaver & Harmy, 1973), and can contaminate cytoplasmic preparations to varying extents, depending on the experimental material. Damage to ribosomes, both structural and by the removal of associated proteins, can occur during isolation and preparation. The extent of this damage can be assessed, to some extent, by extracting the RNA and fractionating it by polyacrylamide gel electrophoresis (see Leaver & Key, 1970).
10.3—
The Biochemical Mechanism of Protein Synthesis
Protein synthesis, i.e. the assembly of polypeptide chains from their amino acid constituents, occurs on the ribosomes. The information specifying the amino acid sequence of the polypeptide chain is contained in mRNA molecules, which
in the case of eukaryotes are complexed with specific proteins in vivo. Messenger-like RNA-protein particles called 'informosomes' have been isolated from the cytoplasm and from polysomes of germinating wheat embryos (Ajtkhozhin et al., 1973; Ajtkhozhin & Akhanov, 1974). So far, however, the RNA of these particles has not been definitely established as mRNA. When an mRNA molecule associates with ribosomes, the information it contains is translated; the number of ribosomes involved depends largely upon the size of the polypeptide chain being synthesized, since generally, ribosomes are evenly spaced along mRNA molecules.
The translation process can be arbitrarily split into initiation, elongation, and termination and release. However, prior to the initiation of the process on the ribosome, the twenty different protein amino acids must be activated and attached to specific tRNA molecules. Also, after release of the polypeptide chain from the ribosome, a series of post-translational changes may occur.
10.3.1—
Amino Acid Activation and Aminoacyl-tRNA Synthesis
This energy-requiring process is accomplished in the cytosol by enzymes called aminoacyl-tRNA synthetases or ligases. It occurs as a two-stage reaction, both stages being catalysed by the same aminoacyl-tRNA synthetase enzyme (Fig. 10.4). The product of the reaction, aminoacyl-tRNA molecules, contain in the
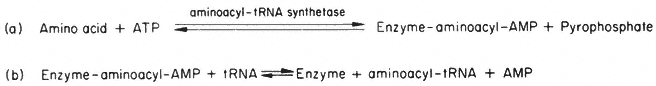
Figure 10.4
The reactions of aminoacyl-tRNA synthesis. (In the cell a pyrophosphatase
acts on the pyrophosphate formed to make reaction (a) irreversible in practice).
aminoacyl-tRNA bond, sufficient energy for the subsequent formation of peptide bonds to occur spontaneously and, in addition, the anticodon triplet of bases of the tRNA ensures that the amino acid will be located at a particular residue position in the amino acid sequence of the protein product. This reaction, therefore, controls the proportion of free to aminoacylated-tRNAs in the cytoplasm, and theoretically therefore, could affect the rate and/or type of protein being synthesized.
10.3.1.1—
tRNA
The code words (codons) of the genetic code were first established for E. coil. This work showed that there were 61 codons shared between twenty protein amino acids, i.e. there is more than one codon for some amino acids and these can attach to more than one tRNA species. Chemically different tRNA species which can be acylated by the same amino acid are called isoacceptor tRNAs.
However, it was soon realized that not every codon has a corresponding tRNA with a specific anticodon and that some tRNAs recognize more than one codon. Crick's 'wobble hypothesis' (1966) to account for this, proposed alternative base-pairing to occur between the base in the third position of the mRNA codon and the corresponding base in the tRNA anticodon. The proposed rules are set out in Fig. 10.5. Thus, inosine in the 5' position in the anticodon of an

Figure 10.5
The basis of the 'wobble' hypothesis. (X =
Any nucleotide; I = Inosine; G = Guanine;
U = Uridine; A = Adenine; C = Cytosine;)
(From Boulter et al. (Biol. Rev. 47, 113–75, 1972.))
aminoacyl-tRNA molecule allows this base to pair with either U, C or A in the third 3' position of a messenger codon. Similarly, with G or U in the 5' position in the anticodon, two codons may be recognized by a single aminoacyl-tRNA, whereas with C or A only a single codon is recognized. Since alternative base-pairing is only possible with the base in the third position of the codon, codons for the same amino acid which differ in either of the first two base positions, base-pair with different tRNAs. The only example of 'wobble' in the first base of a codon occurs with tRNAiMet , which recognizes both the initiator AUG or GUG codons (see later). Even so, separation of tRNAs by counter current-distribution, MAK columns, BD-cellulose columns and reversed phase chromatography has shown that there are more isoacceptor tRNAs than can be accounted for by the degeneracy of the genetic code. Similarly, isoacceptor tRNAs for amino acids exist in plants; for example, soyabean seedlings have at least six different tRNALeu species (Anderson & Cherry, 1969). Although few experiments have been carried out, it has been found in every case investigated that plants use the same codon in amino acid assignments as those of E. coli. However, plants may not use all of the possible codons for a particular amino acid, since some isoacceptor tRNA species may be absent, or because of the 'wobble' effect; e.g. Caskey et al., (1968) have shown that the isoleucyl tRNA from guinea pig liver recognizes AUU, AUC and AUA codons, whereas the corresponding isoleucyl-tRNA from E. coli only recognizes two of these, AUU and AUC. This response to different codons in the two organisms is explained if the liver tRNA had 5' inosine in the anticodon, while E. coli tRNA had guanine.
The whole question of the number and the activity of tRNAs in different tissues of plants is complicated by the fact that, (a) it is not certain whether the methods for separating tRNAs are completely effective; (b) it is known that during isolation and purification, partial modification of tRNA molecules can occur, which may affect their charging ability with amino acids or their ability to transfer amino acids to proteins, or both (see Sueoka & Kano-Sueoka, 1970); (c) the relative importance of the three protein synthesizing systems, cytoplasmic, chloroplast and mitochondrial, each with its associated tRNAs and synthetases may differ in different tissues. Nevertheless, different complements of isoacceptor tRNAs may occur in different tissues and changes in pattern have been demonstrated during development (Bick et al., 1970; Littauer & Inouye, 1973), and it has been suggested that isoacceptor tRNAs are involved in the control of protein synthesis. Garel et al., (1973) have shown that the complement of tRNAs synthesized in the silk gland is related to the mRNAs being translated and they have suggested defining this phenomenon as 'modulated tRNA biosynthesis'.
10.3.1.2—
Aminoacyl-tRNA Synthetases
Aminoacyl-tRNA synthetases have been isolated and purified from a variety of plants (see Boulter 1970; Boulter et al., 1972; Zalik & Jones, 1973), and although a considerable amount of work has been done on their properties, several problems remain. For example, the question of the detailed substrate specificity of different aminoacyl-tRNA synthetases is still under investigation. Since activation is a two-step process, both amino acid and tRNA specificity have to be considered. The synthetase is normally absolutely specific towards the tRNA; few mismatches have been reported in homologous systems (Arca et al., 1967, 1968). However, a synthetase specific for one amino acid can, in some instances, activate to a lesser extent another amino acid. Even so, synthetases are remarkably amino acid specific. A second amino acid attached to a tRNA molecule normally specific for another amino acid, will be located in the polypeptide chain according to the tRNA anticodon, i.e. where the first amino acid would have been placed. With regard to amino acids not normally found in the cell, specificity is not always so pronounced, e.g. azetidine-2-carboxylic acid is activated and transferred to tRNAPro by the proline enzyme of mung bean (Peterson & Fowden, 1965), whereas Polygonatum multiflorum, which contains a high level of this amino acid in nature, contains a prolyl-tRNA synthetase which will not activate it. In Mimosa and Leucaena, where mimosine, an analogue of phenylalanine, occurs naturally and where it is non-toxic, the phenylalanyl-tRNA synthetase discriminates against mimosine. In several other species where mimosine is toxic, it is activated by the phenylalanine-tRNA synthetase but the enzyme does not attach it to tRNAPhe (Smith & Fowden, 1968).
It is not clear if there is one or more than one synthetase whenever there are
a number of isoacceptor tRNAs. In soybean seedlings where there are at least three different leucyl aminoacyl-tRNA synthetases (Kanabus & Cherry, 1971) which have different specificities towards the six leucine isoacceptor tRNAs, it would appear that there is more than one cytoplasmic leucyl-tRNA synthetase. However, in some instances, a single aminoacyl-tRNA synthetase can recognize different isoacceptor tRNAs. When both cytoplasmic and chloroplast tRNA species are present, there are probably at least two sets of aminoacyl-tRNA synthetases. In light-grown Euglena for example (Reger et al., 1970; Krauspe & Parthier, 1973), there are two aminoacyl-tRNA synthetases for each of phenylalanine and leucine, whereas dark-grown Euglena, which is without chloroplasts, contains only one; there are various reports of chloroplast aminoacyl-tRNA synthetases which only acylate chloroplast tRNAs and not their cytoplasmic counterparts and vice versa. However, in heterologous systems such as chloroplast tRNA plus cytoplasmic activating enzymes, specificity may range from none to complete (Burkard et al., 1970; Boulter et al., 1972), although the biological significance of these findings is not clear.
There is evidence that, as well as the tRNAs, the complement of aminoacyltRNA synthetases may change during the life cycle of plants (Bick & Strehler, 1971).
10.3.2—
Translation of mRNA
10.3.2.1—
Initiation of the Polypeptide Chain
Initiation of protein synthesis on plant cytoplasmic ribosomes starts with the assembly of a mRNA•40s subunit complex which requires ATP for its formation and this step is followed by the addition of methionyl-tRNAi to form a mRNA•40s subunit•methionyl-tRNAi complex. The process requires GTP and at least two initiation factors (Weeks et al., 1972; Marcus et al., 1973). Initiation is completed by the addition of the 60s subunit and the 80s ribosome so formed is then able to accept the next aminoacyl-tRNA, thus starting the elongation process. The tRNAi Met is a special tRNA charged with unformylated methionine, which can enter the so-called 'P' site of the ribosome complex and base-pair with the initiator codon, AUG or GUG in the mRNA. Although mRNAs are translated from the 5'® 3' end, evidence from sequence studies of bacteriophage messages shows that the initiator triplet is not at the 5' terminus, but is set a considerable number of nucleotides in. For example, the first initiator codon of bacteriophage R17 RNA is preceded by 91 nucleotides which are not translated (Adams & Cory, 1970). In the case of polycistronic messengers, one or more initiator triplets will occur intramolecularly.
Evidence that plants have a special type of initiator tRNA, tRNAi Met , comes from the wheatgerm system (Leis & Keller, 1970; Marcus et al., 1970a; Tarrago et al., 1970; Ghosh et al., 1971), and from Vicia faba (Yarwood et al., 1971a),
where it has been shown that two major and one minor tRNAMet species are present. The minor and one of the major tRNAMet species function as chain initiators as shown by AUG-dependent binding (Tarrago et al., 1970; Yarwood et al., 1971a), and by N-terminal analyses of either the product of tobacco mosaic viral RNA-directed (Marcus et al., 1970a) or poly-AUG, poly-GU and endogenous messenger-directed incorporation (Yarwood et al., 1971a). Neither of the major tRNAMet species from either wheat or beans is formylated, in contrast to the initiator tRNAiMet of prokaryotes, and it is presumed that these are the cytoplasmic tRNAMet s for initiator and internal methionine residues. The minor, formylatable tRNAMet , is presumed to be the initiator of chloroplast protein synthesis (see chapter 11)
The hydrolysis of ATP precedes the formation of the first peptide bond and may be required for recycling of the methionyl-tRNAi binding factor. The requirement for ATP has also been demonstrated in the rabbit globin synthesizing system (Schreier & Staehelin, 1973), but not in those of prokaryotes. The order in which the initiator tRNA and mRNA bind to the small subunit in mammalian and prokaryote systems is in dispute; the order suggested above for wheatgerm by Marcus and his coworkers, therefore, whilst agreeing with some workers on those systems disagrees in particular with the suggestion of Legon et al., (1973) and Schreier and Staehelin (1973) that the initiator binds prior to the mRNA in rabbit globin synthesis.
Knowledge of the two plant initiator factors involved in binding methionyl-tRNAi and mRNA is much less complete than for the similar factors in bacterial and mammalian systems; details of the latter are, therefore, relevant. In bacteria there are three factors, IF-1, IF-2 and IF-3 involved in the initiation process. Factor IF-1 has a molecular weight of 9,400 daltons and is the most basic of the three proteins. Factor IF-2 has been shown to consist of two sub-components, both active, with molecular weights of 9 x 103 –100 × 103 daltons and 80 × 103 daltons although the latter is probably derived from the former by proteolysis during purification. Factor IF-3 is a protein or group of proteins with molecular weight(s) of 21.5 × 103 –23.5 × 103 daltons (Grunberg-Manago et al., 1973). The three factors attach to the small subunit; IF-2 binds GTP and formyl-methionyl-RNA, while IF-3 binds mRNA. Factor IF-I increases the affinity of factors IF-2 and IF-3 for the 30s subunit, and is also necessary for the release of IF-2 from the ribosome.
The order of the attachment of the different factors to the small subunit and the order of the binding of mRNA and acylated initiator tRNA, is still a matter of debate. Some results suggest that IF-1 is attached before IF-2 and IF-3, whilst others indicate that it can only be attached after. Similarly, it is possible that an unstable intermediary complex consisting of formyl-methionyl-tRNAi •30s subunit•initiation factors is formed, i.e. that mRNA is bound after initiator-tRNA, or that a 30s subunit•mRNA•IF-3 complex is formed, which is then stabilized by the addition of formyl-methionyl-tRNAi and IF- I and IF-2. The next step in the process, the addition of the large subunit, does not require GTP, but
GTP hydrolysis is required to release IF-2 from the 70s ribosome complex. The role of GTP hydrolysis in initiation is still not fully understood, but it is not only required for the release of IF-2. Factor IF-3 is not only required for the binding of natural mRNAs, but also functions later either to dissociate the subunits after chain termination, or to keep dissociated subunits apart until another cycle of initiation is instituted. This activity of the IF-3 factor is referred to as its DF (ribosome dissociation factor) activity. None of the factors occurs on the polysomes and they recycle in protein synthesis, as shown in Fig. 10.6.
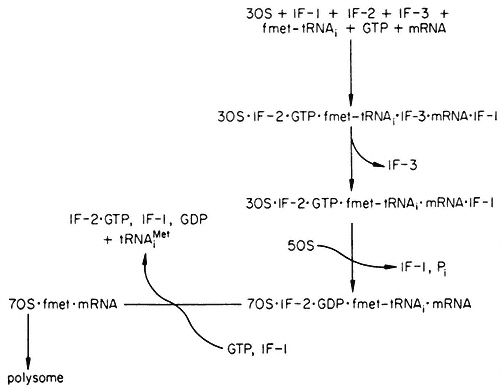
Figure 10.6
Recycling scheme for initiation factors in prokaryotes. The exact order
in which the factors interact with the 30s subunit is still not clear, nor is
whether fmet-tRNAi attaches before or after mRNA. IF-2•GTP•fmet-
tRNAi may exist independent of the 30s subunit. (fmet-tRNAi =acylated
formylmethionyl-tRNAi ; tRNAiMet =deacylated formylmethionyl-tRNAi ).
(Modified from Haselkorn & Rothman-Denes (1973).)
Several proteins which form an integral part of the small subunit have also been shown to be involved in initiation. Small subunit proteins, s1, s11, s13, s19, participate in IF-2 binding and s1, s11, s12, s13, s14, in IF-3 binding; s12 is responsible for recognition of mRNA (Haselkorn & Rothman-Denes, 1973; Anderson et al., 1974). Shine and Dalgarno (1974) have proposed a model of how the large subunit.initiation factor complex may recognize and attach mRNA. It would appear that the sequence (5') GGAGGU (3') is present in the same relative position with respect to the first translatable AUG triplet in all prokaryotic messengers so far analyzed. Furthermore, the 3' proximal end of 16s RNA has the sequence GAUCACCUCCU UA (OH), so that the underlined nucleotides could potentially base-pair with the GGAGGU sequence in the mRNA.
Further complexity of the initiation process is indicated by the work of
Schreier and Staehelin (1973), who have characterized five initiation factors (IF-E1 to IF-E5 ) for mammalian protein synthesis. Their results with rabbit globin mRNA are consistent with the formation initially of a methionyl-tRNA1 •IF-E2 •GTP complex, which is bound independently of mRNA to the 40s ribosome subunit by IF-E3 . Subsequent to this, mRNA and the 60s subunit are joined by the cooperative action of IF-E4 , ATP, IF-E1 and IF-E5 . The binding of natural mRNA requires IF-E4 and ATP. IF-E5 promotes the joining of the 40s complex with the 60s subunit and IF-E1 inhibits complex formation in the absence of mRNA binding. However, it has been suggested that the apparent need for additional factors results from deproteinization of ribosomal subunits and mRNAs, i.e. the requirement may be for structural proteins rather than initiation factors. The probable relationship between the different factors 1s given in Table 10.1. The little information available suggests that eukaryotic factors are not exchangeable with their prokaryotic counterparts.
| ||||||||||||||||||||||||||||||||||||||||
Factor EIF-3, contrary to its prokaryotic counterpart, consists of a number of different sized polypeptide chains; the question as to whether there are different EIF-3s or whether selectivity is controlled by additional mRNA specific factors, is still unresolved. A similar uncertainty to that described for the prokaryotic system surrounds the question as to whether or not methionyl-tRNAi binds to the 40s subunit prior to mRNA.
The initiation process is extremely complex and still not fully understood. A variety of proteins are involved, some being ribosomal structural proteins and others proteins which recycle during the process. These proteins interact, changing the conformation of the ribosome and thereby allowing the different steps of initiation to proceed in a sequential and orderly manner. Different mRNAs probably differ in their rate of attachment to the ribosome and/or in their efficiency in other stages of the initiation process, so affecting the frequency and rate of translation. With proteins which contain prosthetic groups their
absence may inhibit initiation, since it has been shown that lack of haemin in globin synthesis, results in the formation of an inhibitor of the binding of methionyl-tRNAi to 40s subunits (Adamson et al., 1972; Gross & Rabinovitz, 1973; Legon et al., 1973). Furthermore, a variety of mRNA specific factors have been proposed called 'interference' or 'i-factors', which affect the specificity of the initiation factor IF-3 (Groner et al., 1972). Thus, Strycharz et al. (1973) have identified a supernatant factor in Krebs II ascites cells, which is specifically required for the initiation of the translation of encephalomyocorditis viral RNA. However, Lodish (1974), from a kinetic analysis of protein synthesis, has proposed a model which precludes the necessity for mRNA specific factors. He points out that these have often been observed in systems which are less active than the corresponding in vivo system, and that the three eukaryotic systems available which translate exogenous mRNAs efficiently in vitro, do so with a variety of mRNAs.
10.3.2.2—
Elongation of the Polypeptide Chain
The elongation process starts once the 80s plant ribosome containing mRNA and methionyl-tRNAi , has been assembled and takes place by the following repeated cycle of events, each cycle being separated into:
(a) codon-directed binding of aminoacyl-tRNA;
(b) peptide bond formation; and
(c) translocation.
The ribosome has two binding sites; a 'P' site for the binding of methionyl-tRNAi and an 'A' site where all other aminoacyl-tRNAs bind. After the initiation complex has been formed, the next aminoacyl-tRNA, i.e. the one carrying the anticodon to the next codon of the mRNA, binds at the 'A' site. Once the aminoacyl-tRNA has been bound in the 'A' site, a peptide bond is formed by a peptidyl-transferase enzyme, which is part of the large subunit, such that the 'A' site now carries the aminoacyl-tRNA joined to methionine by a peptide bond. The 'P' site now carries the deacylated tRNAiMet , which is subsequently removed leaving an open 'P' site, and the peptidyl-tRNA moves from the 'A' to the 'P' site, so moving the message relative to the ribosome. The 'A' site is now empty and is ready for the next aminoacyl-tRNA to enter according to the next codon in the message. This cycle of reactions requires K+ , Mg2+ , GTP and various elongation factors, proteins, which are found in the soluble fraction of cell homogenates.
Our knowledge of polypeptide chain elongation is most complete with E. coli, where three elongation factors are known, EF-Tu, EF-Ts and EF-G (Fig. 10.7). Factor EF-Tu binds stepwise with GTP and aminoacyl-tRNA and this ternary complex transfers the aminoacyl-tRNA to the 'A' site of the ribosome, releasing an EF-Tu•GDP complex and inorganic phosphate. Factor EF-Ts displaces GDP to form the complex EF-Ts•EF-Tu, from which GTP displaces EF-Ts to regenerate EF-Tu•GTP. The third elongation factor, EF-G,
and free GTP, are responsible for the translocation of the peptidyl tRNA from the A' to the 'P' site with the prior removal of the deacylated tRNA from the 'P' site; GTP is hydrolyzed during the process. The elongation factors, like the initiation factors, recycle during protein synthesis (Fig, 10.7). The protein chain grows from the N-terminal end, and probably starts to fold into its three-dimensional conformation whilst the process of elongation is proceeding.
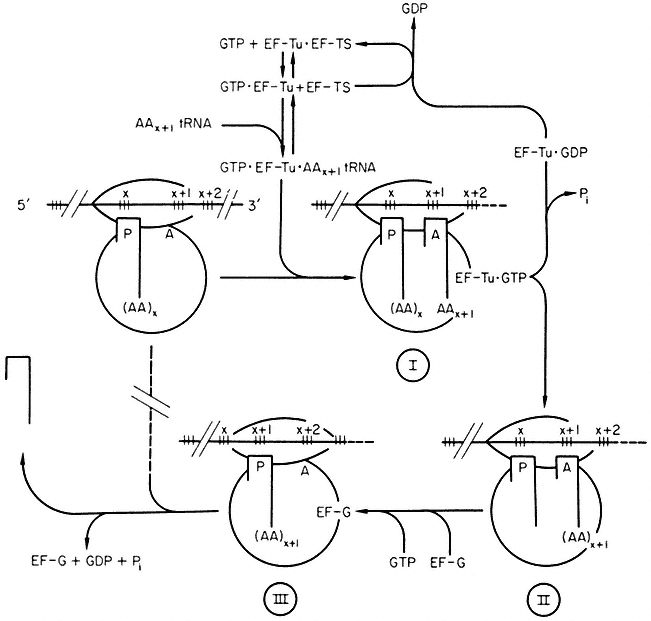
Figure 10.7
Polypeptide chain elongation in prokaryotes. Ribosomes cover more
than two codons on mRNA. Reaction I ® II is catalysed by peptidyl transferase.
(
(Modified from Haselkorn & Rothman-Denes (1973).)
Fewer details of the elongation factors are available in eukaryotic systems. However, two elongation factors, designated as the binding enzyme EF-1 and the translocase EF-2 (Hardesty et al., 1963; Gasior & Moldave, 1965) have been purified from ungerminated wheat embryos (Golinska & Legocki 1973; Twardowski & Legocki, 1973; Legocki, 1973; Allende et al., 1973). The mechanism of binding of aminoacyl-tRNA to wheat ribosomes is on the whole similar to
that in E. coli, the main difference being the absence in the eukaryotic system of a factor with the properties of EF-Ts, i.e. EF-1 = EF-Tu and EF-2 = EF-G.
Elongation factor EF-I, which requires the presence of Mg2+ at low concentration, K+ and GTP, is responsible for the binding of aminoacyl-tRNA to wheat ribosomes. It contains three active forms differing in molecular weight. The larger EF-1 forms dissociate to a single species of molecular weight 6 × 104 daltons during gel electrophoresis in the presence of SDS. The three forms observed may represent a monomer, trimer and tetramer of a single protein unit, of which the trimer seems to be the most stable form. Some experimental evidence supports the view that the light species is the active form of the EF-1 enzyme. For example, in calf brain and wheat embryo, it has been shown that the interaction of EF-1 with GTP and aminoacyl-tRNA yields a ternary complex which contains the light form of the enzyme, i.e. molecular weight 5 × 104 to 6 × 104 daltons (Moon et al., 1972; Legocki, 1973). It is suggested that the amino group of the esterified amino acid plays an important role in aminoacyl-tRNA binding (Jerez et al., 1969) and this differs from the bacterial system where EF-Tu can react with the deaminated product of phenylalanyl-tRNA, phenyl-lactyl-tRNA (Fahnestock et al., 1972).
The second elongation factor, EF-2, is involved in translocation of the peptidyl-tRNA from the 'A' site to the 'P' site during elongation. It is a protein of molecular weight 7 × 104 daltons and requires thiol compounds for activity (Twardowski & Legocki, 1973).
Ribosomes and elongation factors from the cytoplasm of eukaryotic organisms are not functionally interchangeable with their E. coli counterparts (Krisko et al., 1969). Furthermore, Perani et al., (1971) have shown absolute ribosome specificity, either for 70s or 80s ribosomes, of two sets of elongation factors, T and G (EF-1 and EF-2), isolated from yeast, and for factor EF-2 isolated from the alga, Prototheca zopfii. It is generally accepted that elongation factors within each ribosomal type (70s or 80s) are exchangeable between eukaryotes (Ciferri, 1972).
10.3.2.3—
Termination and Release of the Polypeptide Chain
No reliable information is available on chain termination in plants, most of the work having been done with E. coli. However, as there are some differences between the prokaryote and mammalian eukaryote systems, these will be mentioned briefly. In E. coli the elongation cycle is repeated until certain termination codons on the mRNA come into the 'A' site; these codons are UAG, UAA and UGA. At least three release factors, proteins, which recognize the terminator codons and bring about the release of the completed polypeptide chain from the tRNA, have been identified (Caskey et al., 1969; Capecchi & Klein, 1969; Milman et al., 1969). Originally designated R-1, R-2 and S these are now called RF-1, RF-2 and RF-3. RF-1 recognizes UGG or UAG, RF-2
recognizes UAA or UGA, and RF-3 affects the rate of release of the polypeptide chain. In mammalian systems, only a single release factor which responds to all three codons has been identified, and furthermore the prokaryotic termination requirement for GTP appears to be absent (Haselkorn & Rothman-Denes, 1973).
In addition, several other termination factors have been proposed from different organisms (see Haselkorn & Rothman-Denes, 1973). Although little is known about chain termination in plants, the presumed universality of the code suggests that similar terminator triplets and proteins are involved in plants also.
Originally, it was thought that on chain termination ribosomes dissociated into subunits and that the free ribosomes found in the cell were inactive in further protein synthesis. Noll et al. (1973) have presented evidence that at least some of the free ribosomes found in the cell interact with a factor and partially dissociate, so that the small subunit with bound IF-1, IF-2 and IF-3, can initiate protein synthesis.
In conclusion, it may be said that small differences exist between the mechanism of protein synthesis in the pro- and eukaryotic systems, e.g. the initiator tRNA, the involvement of GTP in different steps, as well as in the possible number of elongation factors. In view of the technical difficulties involved, work on the detailed mechanisms of eukaryotic systems could be considered hardly worthwhile. However, it is possible that translation level controls are much more important in eukaryotes than they are in prokaryotes. This possibility, together with the importance of plant proteins as a source of food, are justification enough for an attempt to characterize the process in detail in plants and, thereby develop suitable assay systems.
10.3.2.4—
Cell-Free Systems
In order to gain detailed knowledge of the mechanism of protein synthesis in plants, it is necessary to develop in vitro assay systems. Such systems must satisfy the following criteria:
(a) be dependent on the exogenous component(s) to be tested;
(b) be efficient, i.e. of comparable activity to in vivo rates, in order to allow quantitative analysis;
(c) the product, whose synthesis is monitored, must be clearly defined.
Two basic types of cell-free systems are used: fractionated systems, in which enzymes and components are isolated, purified and then reconstituted into an assay system, and unfractionated systems, in which the cells are broken open and cell-debris, nuclei and mitochondria removed by centrifugation leaving the ribosomes and the various components for protein synthesis in the supernatant, which is then used as the assay system. If all of the components of the assay are isolated from one organism, the system is said to be homologous as opposed to heterologous when they are not.
In addition to the fractionated E. coli prokaryotic system, there are four
fractionated eukaryotic systems which satisfy the above criteria. These are the rabbit reticulocyte (Lockard & Lingrel, 1969; Gilbert & Anderson, 1970), the mammalian liver (Prichard et al., 1971), the Krebs II ascites cell (Mathews & Korner, 1970), and the only plant system, that of wheatgerm (Allende & Bravo, 1966; Allende, 1970; Leis & Keller, 1970; Marcus et al., 1970a,b; Tarrago et al., 1970; Ghosh et al., 1971; Legocki & Marcus, 1970; Klein et al., 1972; Lundquist et al., 1972). Other fractionated cell-free systems from plants, such as those from developing legume seeds (Gumilevskaya et al., 1971; Payne et al., 1971a,b; Yarwood et al., 1971a,b; Beevers & Poulson, 1972; Wells & Beevers, 1973), are not well characterized and assays often lack quantitative accuracy. This is because in isolating the constituent enzymes and components, damage is caused in breaking the cell wall by hydrodynamic sheer, by cell vacuoles breaking and releasing acids, phenolics, tannins and other substances, and by activation of proteases and nucleases (Payne & Boulter, 1974). If the preparations used contain several components and structural damage to ribosomes has occurred, interpretation of the results may be qualitatively ambiguous, since enzyme preparations may contain proteins needed for the reconstitution of protein-leached ribosomes.
Unfractionated cell-free systems cannot be made from tissues which have a high nuclease or protease activity, and until better methods for the inhibition of degradative enzymes become available, most unfractionated systems from plants will be of limited value. Recently, Davies et al. (1972) have used media of high ionic strength and high pH with some success, and Gray and Kekwick (1973) have developed a system from pea seedlings using 0.2 mM vanadyl sulphate as an inhibitor. This system has been shown to synthesize the small subunit of ribulose bisphosphate carboxylase (Fraction 1 protein), since the tryptic peptides of the in vitro product were similar to those of the naturally occurring protein. The identity of the product was also proved by immunoprecipitation. The requirement of product identification is essential in complete cell-free assays (see also chapter 11, where the synthesis of the large subunit of Fraction I protein by the chloroplast cell-free system is described).
The wheatgerm system is by far the most successful unfractionated cell-free system from plants. Its preparation is as follows (Marcus et al., 1974):
Dry wheat embryos are ground thoroughly with a small amount of sand in a precooled mortar in a total volume of 3.3 ml of 90 mM KC1, 2 mM CaCl2 , 1 mM Mg (Ac)2 , 6 mM KHCO3 . The embryos are initially ground in 1.0 ml with 0.5 and 1.8 ml increments added subsequently. The slurry is then centrifuged for 10 minutes at 23,500 × g and the supernatant is removed with a Pasteur pipette, taking care to leave behind as much as possible of the upper lipid layer. Just prior to use, 0.5–3.0 ml are dialyzed against 500 ml of 1 mM Tris–acetate, pH 7.6, 50 mM KCl, 2 mM Mg(Ac)2 , 4 mM 2-mercapto-ethanol for 1.75 hours; this preparation is termed S23.
The wheatgerm system is attractive in spite of needing some additions, (e.g. tRNA), since it shows great promise as a 'translation' system for various mRNAs. This is an important development as the isolation of mRNAs from plants is now feasible using binding to oligo-dT columns (since a proportion of eukaryote messengers contain a poly-A sequence), by gradient centrifugation and by immunoprecipitation of polysomes (Haselkorn & Rothman-Denes, 1973; see also chapter 9).
Four RNAs of brome mosaic virus (BMW) induce amino acid incorporation into proteins when used as messengers in the wheatgerm system. RNA4 is translated with an efficiency comparable to that of bacteriophage RNA in E. coli extracts. The product is homogeneous and indistinguishable from the coat protein of BMV (Shih & Kaesberg, 1973). Satellite tobacco necrosis virus RNA has also been translated in this system and the product shown to be coat protein (Klein et al., 1972). Several natural eukaryotic messengers have also been translated in the wheatgerm system, e.g. rabbit globin mRNA (Efron & Marcus, 1973) and leghaemoglobin RNA (Verma et al., 1974). The latter is the only naturally occurring plant messenger to be isolated so far. It is a poly A-containing 9s–12s mRNA which was isolated from soybean root nodule polysomes. When used to programme the wheatgerm system by Verma et al., (1974), the product was identified serologically as mainly leghaemoglobin S (LbS ) with a little leghaemoglobin F (LbF ); there are two types of leghaemoglobin found in soybean nodules and the reason for the preferential synthesis of the LbSin vitro, is not understood. Factors other than those already present in the wheatgerm system were not required, and there was no need for the addition of heme.
There are several unfractionated cell-free systems from animals (Haselkorn & Rothman-Denes, 1973). Of particular interest is that of the Xenopus oocyte since intact eggs are used to translate exogenous messages (Gurdon et al., 1971). Natural plant messages have not been used but there is evidence that tobacco mosaic virus RNA can be translated (Knowland, 1973). Coat protein is not synthesized, however, and the main product is a polypeptide with molecular weight 14 × 104 daltons. Although no function has been assigned to this protein, it is known that plant cells infected with TMV also make a protein of the same molecular weight (J. Gurdon, personal communication).
The use of heterologous systems assumes the interchangeability of the various enzymes and components between different systems and as mentioned previously, it is generally accepted that this is not usually possible between prokaryotic and eukaryotic organisms. Generally, attempts to demonstrate interchangeability between different eukaryotic systems have been positive, but several workers have suggested that this is because discrimination can only be seen under optimum conditions. The general conclusion is that tissue and organism specificity may occur but only in exceptional cases (Mathews, 1973).
10.3.2.5—
Inhibitors
Several antibiotics and other inhibitors block various steps in protein synthesis, and are, therefore, extremely useful in establishing the mechanism of the process (Pestka, 1974). Table 10.2 lists the more common inhibitors and their sites of action; many of these have been used with plants. It is important to realize
| ||||||||||||
that the concentration of the inhibitor is often critical for the production of a specific effect; it is essential to establish the conditions for the correct use of an inhibitor with each tissue and to maintain effective controls. It has been suggested by Glazer and Sartorelli (1972) that in rat liver, membrane-bound 80s ribosomes are more susceptible to a range of inhibitors than are the 80s ribosomes free in the cytosol.
10.3.3—
Post-Translational Changes
Changes which take place after the synthesis of the polypeptide chain are called post-translational. The most common of these are concerned with the many proteins which consist of more than one polypeptide chain. Since there is no evidence that unpartnered subunits occur in any quantity, machinery must exist in the cell to ensure that approximately correct numbers of each constituent polypeptide chain are synthesized and correctly assembled, although little is known about the mechanisms. Some mRNAs may be polycistronic but in other
cases mRNAs for each polypeptide chain are translated independently. Self-assembly by interaction at specific sites on different polypeptide chains may occur.
Several examples are known where some subunits of a protein are made in the cytoplasm and others on the chloroplast or mitochondria. For example, ribulose bisphosphate carboxylase is a large molecule (also known as Fraction I protein) consisting of two types of subunit; the small subunits are synthesized on 80s cytoplasmic ribosomes, whilst the large subunits are synthesized on 70s chloroplast ribosomes and assembly of the two types to form the complete molecule occurs in the chloroplast (see chapter 11).
Post-translational changes also occur by the addition of prosthetic groups or by modification of the polypeptide chain. Such modifications may be the formation of disulphide linkages, the removal of N-terminal portions, the removal of intramolecular segments, the derivation of amino acids and the addition of carbohydrate or lipid units to form glycoproteins and lipoproteins respectively. These changes are all brought about by specific enzymes and probably after the polypeptide chain has left the ribosome.
The formation of disulphide linkages is often part of the assembly of multichain proteins, e.g. legumin in Vicia faba (Wright & Boulter, 1974), although the enzymes involved have not been demonstrated from plants. Few plant proteins have methionine at their N-terminus when isolated, although this amino acid must be presumed to be the N-terminus when synthesized, and, therefore, removal of one or more residues at the N-terminal region must have occurred subsequently. Enzymes for this purpose have been demonstrated in bacteria but not in plants, and it is possible that the process in plants is non-specific and mediated by leucine aminopeptidase, a ubiquitous plant enzyme which removes amino acids from the N-terminus of proteins and may continue to do so until the conformation of the protein inhibits its activity. The removal of intramolecular segments of plant proteins similar to the situation with proinsulin in animals, has not been demonstrated, but there is no reason to suppose that it does not occur. Modification of individual amino acids is quite common in plant proteins; examples are, S -N-trimethyllysine in some cytochromes c (Ramshaw et al., 1974), and hydroxyproline in extensin, a protein of cell walls (Lamport, 1965). Where investigated, the protein when synthesized contained the parent amino acid, the substituted amino acid arising secondarily, probably after the protein had left the ribosome. Some residue positions of a particular amino acid may be substituted and others not, though the specificity of the changes is not understood.
Synthesis of the protein moiety of glycoproteins takes place on the membrane-bound ribosomes of the ER, but the sugar groups are added sequentially at different sites in the cell. Those closely linked to the polypeptide chain such as glucosamine and mannose, are added immediately after release from the ribosome or in the ER itself, other more terminal groups may be added later, some in the Golgi apparatus (Whaley et al., 1972).
10.4—
Protein Synthesis in vivo
The rate of protein synthesis in vivo can be measured by determining the amount of protein in a plant, organ or tissue at two different times; this will give the net rate since proteins 'turnover' and the amount of protein found would represent the balance between synthesis and degradation (Huffaker & Petersen, 1974).
Various equations describing the relationship between the synthesis and degradation of protein have been proposed but there are many technical and interpretive difficulties in this type of experiment. The turnover rate for an individual protein is not the same as the value obtained for the total protein and although the latter may be of interest agriculturally, most biochemical and physiological studies require information on individual proteins. This information has only been obtained so far with a few plant proteins, although most are thought to turnover (Huffaker & Petersen, 1974; see also chapter 12). Possible exceptions are storage proteins which are sequestered from the metabolically active cytoplasm and stored in membrane-bound protein bodies until subsequently required at a more active phase of the life-cycle.
A variety of studies using acrylamide gel electrophoresis has shown that the protein complement of organs and tissues changes during the life-cycle of a plant, and also in response to environmental conditions. Organ-specific proteins have been demonstrated serologically and often one or a few proteins may predominate in an organ, e.g. ribulose bisphosphate carboxylase, which often accounts for 50% of the protein of leaves. The changing complement of proteins during development is due to changes in the type or rate of protein synthesis and/or degradation. For example, Beevers and Poulson (1972) followed changes in the protein content of pea cotyledons during the period 9–33 days after flowering. Initially, protein content increased gradually, then rapidly between 21–27 days to decline once more as the seed dehydrated and matured. Incorporation experiments with 14 C-leucine indicated that albumin proteins were synthesized early in cotyledon development whilst globulin synthesis predominated with increasing maturity. During the period of rapid protein synthesis, greater amounts and a higher percentage of polysomes to monosomes were extracted than at other stages in the development of the seed. However, unlike in bacteria, where induced enzyme synthesis is widespread and the product is not 'turned over' but diluted out by a rapid rate of cell division, plants have a much slower rate of cell division and protein degradation appears to be a common method for regulating the amount, and hence the activity of proteins.
The relative amounts of different seed proteins synthesized may vary considerably in different varieties and breeding lines of a crop; for example, in maize where the major storage proteins of the seeds are prolamins and glutelins normal maize contains a relatively high proportion of prolamin, whereas in the mutant Opaque 2, the proportion of prolamin is greatly reduced. This is of particular significance since prolamin is very deficient in the essential amino
acid lysine, so that seed meals obtained from Opaque 2 have a higher content of lysine and are consequently more nutritious than those from normal maize.
Protein synthesis is a concerted process involving the biochemical machinery of the cell as a whole, since it consists of a series of co-ordinated reactions, the constituents of which may be synthesized in the nucleus (mRNA, tRNA), the mitochondria (ATP), the cytosol (amino acids and aminoacyl tRNAs), the ribosomes (enzymes). Although changes in the rate of protein synthesis occur, very little is known about the mechanisms involved. Theoretically, control could be exerted at any one of the many constituent reactions, this being the pacemaker reaction for protein synthesis in that particular cell/tissue. However, the control mechanisms must allow the overall process to adjust in response to genetic, physiological and environmental stimuli, so that the rate and/or the type of proteins being synthesized can change.
Further Reading
Allende J.E. (1970) Protein biosynthesis in plant systems. Techniques in Protein Biosynthesis, vol. 2 (eds. P.N. Campbell & J.R. Sargeant), pp. 55–100. Academic Press, London and New York.
Anderson W.F., Bosch L., Gros F., Grunberg-Manago M., Ochoa S., Rich A. & Staehelin Th. (1974) Initiation of protein synthesis in prokaryotic and eukaryotic systems. FEBS Letters48, 1–6.
Boulter D. (1970) Protein synthesis in plants Ann. Rev. Pl. Physiol.21, 91–114.
Boulter D., Ellis R.J. & Yarwood A. (1972) Biochemistry of protein synthesis in plants. Biol. Rev.47, 113–75.
Haselkorn R. & Rothman-Denes L.B. (1973) Protein synthesis. Ann. Rev. Biochem.42, 397–438.
Kurland C.G. (1972) Structure and function of the bacterial ribosomes. Ann. Rev. Biochem.41, 377–408.
Mans R.J. (1967) Protein synthesis in higher plants. Ann. Rev. Pl. Physiol.18, 127–46.
Stewart P.R. & Letham D.S. (Eds.) (1973) The Ribonucleic Acids. Springer-Verlag.
Wittman H.G. (1973) Structure and function of bacterial ribosomes. In: Regulation of Transcription and Translation in Eukaryotes, (eds. E.K.F. Bautz, P. Karlson & H. Kersten), pp. 211–212. 24 Colloquium der Gesellschaft für Biologische Chemie. Springer-Verlag Berlin, Heidelberg, New York.
Zalik S. & Jones B.L. (1973) Protein biosynthesis. Ann. Rev. Pl. Physiol.24, 47–68.
Chapter 11—
The Genetic Information of Organelles and Its Expression
11.1—
Introduction:
The Concept of Organelle Autonomy
The most notable event in biology at the dawn of the present century was the rediscovery of the Mendelian laws of inheritance by de Vries, Correns, and Tschermak. The realization by Sutton that the results of Mendel's experiments with garden peas could be explained in terms of the visible behaviour of chromosomes during meiosis led to the nucleus being regarded as the sole carrier of the hereditary material. As often happens in science, this splendidly simple view did not survive for long. The results of experiments by Baur (1909) and Correns (1909) on the inheritance of plastid defects in variegated plants were difficult to explain on the basis that the genes concerned were located in the nucleus. In certain cases, the inheritance of such defects occurred only through the maternal line, i.e. it was uniparental. In other cases the inheritance was biparental, but did not obey the rules of Mendel. For a time it appeared that, far from being 'nuclear', some aspects of inheritance were 'unclear'. It was Baur who pointed out that these results were explicable on the assumption of the genetic continuity of an extrachromosomal entity located in the plastid. Much more extensive evidence for this concept was later provided by the work of Renner (1929) using the genus Oenothera; he suggested the term 'plastome' to describe the genetic system in the plastid. Maternal inheritance is thus explicable in terms of the lack of plastids in the pollen tube of some species; the plastome of the new generation is consequently derived entirely from the mother.
These studies of variegated plants provided the first firm evidence for the existence of extrachromosomal inheritance. Since then, further evidence has accumulated with examples known in representatives of most major groups of organisms. Today we realize that organelle genetic systems are a fundamental feature of the organization of eukaryotic cells. The most detailed work has concentrated on plastids and mitochondria, but there are indications that genetic determinants may occur in other organelles found in eukaryotic cells, especially centrioles, basal granules and kinetoplasts. There is no evidence that either peroxisomes or glyoxysomes contain any genetic material. In this chapter, the biochemical evidence relating to the concept of the genetic autonomy of plastids and mitochondria only will be considered.
The concept of chloroplast autonomy was founded on the observation that, in the algae, chloroplasts can be seen to divide and to be passed to the new cells
in cell division (Strasburger, 1882; Green, 1964). This cytological evidence led to the view, first proposed by Schimper (1885) and Meyer (1893), that plastids do not arise de novo, but are formed by the division of pre-existing plastids; this view was supported by the genetic experiments of Baur and Correns carried out in the following two decades. The discovery in 1962 that chloroplasts contain both DNA and ribosomes brought the idea of chloroplast autonomy back into vogue, and this notion has so dominated our concepts of chloroplast development in recent years that several attempts have been made to grow isolated chloroplasts in culture. It is now clear that such attempts are ill-founded.
Our current dogmas maintain that for any biological system to be autonomous it must contain four components; (a) DNA to code for its entire structure; (b) DNA polymerase to replicate the DNA; (c) RNA polymerase to transcribe the DNA; (d) protein-synthesizing machinery to translate the messenger RNAs into all the necessary proteins. Intermediary metabolism is not necessary in principle since a supply of small molecules could be taken up from the environment. An extensive literature establishes that both chloroplasts and mitochondria do in fact contain these four components; the properties and functions of these are discussed in the rest of this chapter. It is equally clear that the DNA does not code for all the organelle proteins nor does the protein-synthesizing system make all the organelle proteins. For example, many genes concerned with chloroplast and mitochondrial structure and function are inherited in a Medelian fashion, and are therefore presumed to be located in the nucleus, while there is increasing evidence that the majority of chloroplast and mitochondrial proteins are synthesized on cytoplasmic ribosomes. It is now realized that the demonstration of the cytological continuity of plastids from generation to generation is not sufficient to establish that they replicate independently of nuclear control. How, then, are we to regard the concept of organelle autonomy ?
It is my contention that chloroplasts and mitochondria are not autonomous in any rigorous sense; the term is useful only as a quick way to describe the fact that these organelles contain some genes and make some proteins. We must regard the formation of these organelles as resulting from a complex interplay between the genomes of the organelles and the genome of the nucleus, and the fascination of this subject lies in unravelling the details of this interplay at the molecular level. Much more information is available about the autonomy of chloroplasts than about the autonomy of plant mitochondria; research on mitochondrial autonomy has concentrated almost entirely on animal and fungal cells. This imbalance is reflected in this chapter, which is concerned largely with chloroplast autonomy.
The current state of knowledge can be summarized by saying that, while some information has been accumulated about the properties of chloroplast nucleic acids and protein synthesis, only recently has any hard evidence emerged about their biological function. Developments at the genetic level in higher plants, and the establishment of isolated chloroplast systems which synthesize
specific protein and RNA molecules, promise to provide increasing understanding of the precise functions of chloroplast nucleic acids and protein synthesis.
11.2—
Chloroplast Autonomy
11.2.1—
Chloroplast DNA
11.2.1.1—
Discovery
The existence of DNA in chloroplasts has been established by both cytological and biochemical criteria. Ris and Plaut (1962) provided the first convincing evidence that chloroplasts contain DNA. Electron microscopy of chloroplasts in sections of both algal and higher plant cells revealed areas of low electron density which contained fibrils 2.5–3.0 nm in diameter. These fibrils were not seen if the sections were treated with deoxyribonuclease. There are earlier reports that chloroplasts contain regions which can be stained with the Feulgen reagent for DNA, but in general these reports have not proved reproducible. It is now clear that in most species the concentration of DNA inside the chloroplast is below the limit of detection by the Feulgen method. It can however be detected by autoradiography after exposure of cells to -H-thymidine; the report of Rose et al. (1974) on the distribution of DNA in dividing spinach chloroplasts contains an excellent example of this method. It is important when evaluating such studies to consider the controls that are used. If the incorporated label is all present in DNA, it should be removed by treatment of sections with deoxyribonuclease, but not by treatment with ribonuclease. This control works well for tobacco and spinach leaves and several algae, but not or maize leaves, where deoxyribonuclease fails to remove the label from chloroplasts.
Most studies on chloroplast DNA since 1962 have been carried out using isolated chloroplasts. This approach suffers from the difficulty of distinguishing chloroplast DNA from DNA originating from nuclei, mitochondria, and microorganisms which may contaminate the chloroplast pellet. The resolution of these problems is still not complete, and depends mainly on the differing properties of the DNA from the various sources. There are four characteristic properties of chloroplast DNA that have been used to identify it: namely buoyant density, ease of renaturation, lack of histones, and the absence of 5-methylcytosine. These properties are discussed below. It must be remembered that there is a possibility some chloroplast DNA may not meet these criteria. If, for example, chloroplasts in some species contain a minor DNA component which is a nuclear transcript, contains 5-methylcytosine and renatures poorly, it is doubtful whether present techniques could identify it as chloroplast in location. In one species the problem of contamination by nuclei and micro-
organisms can be entirely avoided; DNA has been identified in chloroplasts isolated from sterile enucleated plants of Acetabularia (Gibor & Izawa, 1963; Baltus & Brachet, 1963).
11.2.1.2—
Buoyant Density
It is now clear that chloroplast DNA from the algae Euglena, Chlamydomonas, and Chlorella has a buoyant density sufficiently different from that of nuclear DNA to allow resolution by analytical ultracentrifugation, but that in many higher plants the densities of the two DNA types are often, but not always, too close to permit this. This information has been hard-won; the story of the way in which the interpretation of band patterns in neutral caesium chloride density gradients has changed since 1963 has been told by Kirk (1971) and Tewari (1971). A summary is given here since it illustrates the very real problem of establishing the identity of chloroplast DNA from higher plants by biochemical methods.
The first report of the chemical characterization of higher plant chloroplast DNA was provided by Kirk (1963). He found that chloroplast DNA from Vicia faba has a GC content (37.4 %) which is slightly but significantly different from that of nuclear DNA (39.4%). In the same year a contrasting report was published by Chun et al., (1963), who found that chloroplast preparations from Spinacia oleracea and Beta vulgaris contained two components of much higher densities than the nuclear DNA as judged by caesium chloride equilibrium density gradient centrifugation. However the bulk of the DNA in these chloroplast preparations had a density similar to that of nuclear DNA; this band was attributed to contamination of the chloroplast pellet by nuclear fragments, and thus the chloroplast DNA was regarded as the two high density components. There followed a spate of papers which supported the claim that, in higher plants, chloroplast DNA has an appreciably higher density than nuclear DNA. This picture changed when re-examination by more rigorous methods supported the earlier view of Kirk, and the higher density components are now attributed to contamination by mitochondria and bacteria.
The present position can be summarized by saying that, in all the higher plants examined, chloroplast DNA had a base composition of 37.5 ± 1% GC and a buoyant density of 1.697 ± 0.001 g cm–3 . Table 11.1 lists some values for the buoyant densities of chloroplast and nuclear DNA which have been agreed by Kirk and Tewari. Nuclear DNA is seen to have a smaller, larger, or similar density to chloroplast DNA from the same plant, depending on the species. The GC contents in Table 11.1 have been calculated from the buoyant densities, but a more accurate method has been devised by Kirk (1967). This method releases the purine bases by gentle acid hydrolysis and separates the adenine and guanine by ion-exchange chromatography.
The moral to be drawn from this story is that the purity of subcellular fractions from higher plant tissues must be established by positive methods if
meaningful interpretations are to be made. Microbial contamination can be reduced by surface sterilization of fresh tissues and by the use of sterile solutions, while mitochondrial contamination can be assayed by succinic dehydrogenase or cytochrome oxidase activities. Nuclear contamination can be reduced by incubation of intact chloroplasts with deoxyribonuclease and phosphodiesterase. The extent of nuclear contamination is difficult to measure, but the ease of renaturation and the absence of 5-methylcytosine serve to distinguish chloroplast from nuclear DNA in all cases so far studied.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
11.2.1.3—
Ease of Renaturation
A more useful criterion than the buoyant density for detecting chloroplast DNA from higher plants is the ease with which it will renature after heat or alkali denaturation. Nuclear DNA renatures only to a slight extent in a few hours,
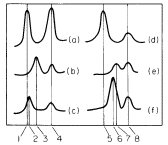
Figure 11.1
Densitometer tracings of ultraviolet absorption photographs of Vicia faba DNA banded
in caesium chloride gradients. (a) Native chloroplast DNA; (b) heat-denatured chloroplast
DNA; (c) renatured chloroplast DNA; (d) native nuclear DNA; (e) heat-denatured nuclear
DNA; (f) renatured nuclear DNA. The numbers refer to densities in g cm-3 as follows: (1)
1.696; (2) 1.699; (3) 1.712; (4) 1.728; (5) 1.696; (6) 1.709; (7) 1.712; (8) 1.728. The
peaks at 1.728 g cm–3 represent marker DNA from Micrococcus radiodurans.
(From Kung & Williams, 1968, by courtesy of Elsevier.)
depending on its content of reiterated sequences. Figure 11.1 illustrates this difference in the case of Vicia faba. The rapid renaturation of chloroplast DNA has encouraged attempts to estimate its genome size from measurements of kinetic complexity. The kinetic complexity of a DNA sample is a measure of the size of the unique set of nucleotide sequences it contains, as judged from the rate at which the DNA renatures. A rapid rate of renaturation implies that like sequences are present in high concentration, and therefore the number of unique sequences, or kinetic complexity, is small. Table 11.2 lists the kinetic complexity, in terms of molecular weight, for the chloroplast DNA from some algae and higher plant species. Some of these values have been corrected from the published figures since the estimate of the molecular weight of the bacteriophage T4 DNA, used as standard, has been revised from 1.3 × 108 to 1.06 × 108 (Dubin et al., 1970). It is striking that the corrected values for the kinetic complexity of chloroplast DNA from the few algae and higher plants examined so far are all in the range 0.9–1.0 × 108 . This may be a coincidence, or it could mean the information content of chloroplast DNA is basically similar throughout the plant kingdom. All the species so far examined belong to the Chlorophyta, with the exception of Euglena. It would be interesting to make such measurements of chloroplast DNA from other plant groups, especially from algae with unusually shaped chloroplasts. Table 11.2 also lists the analytical complexities of chloroplast DNA, i.e. the amount of DNA per chloroplast. Since the kinetic complexities are always much less than the analytical complexities, there must be between 20 and 60 copies of the DNA sequences in each chloroplast. These renaturation studies cannot rule out the possibility of microheterogeneity in nucleotide sequence between the copies but, if it does exist, such heterogeneity is beyond detection by current techniques.
| ||||||||||||||||||||||||||||||||||||
11.2.1.4—
Absence of Histones and 5-Methylcytosine
When released from intact isolated chloroplasts by gentle lysis, chloroplast DNA is not combined with basic proteins. This finding confirms the microscopic observations of Ris and Plaut (1962) that chloroplasts contain DNA fibrils in the same form as they appear in the nucleoplasms of prokaryotic cells. It is also characteristic of chloroplast DNA from both higher plants and algae that it contains no detectable 5-methylcytosine, whereas nuclear DNA invariably does. Whitfeld and Spencer (1968) regard the absence of this base as the most reliable criterion for establishing the purity of chloroplast DNA.
11.2.1.5—
Circularity
A recent discovery is that, when precautions are taken to minimize shearing, a proportion of chloroplast DNA can be isolated as circles. Circular DNA has been reported from chloroplasts of Euglena (Manning et al., 1971), Pisum sativum (Kolodner & Tewari, 1972), Spinacia oleracea (Manning et al., 1972), Antirrhinum majus, Oenothera hookeri, and Beta vulgaris (Hermann et al., 1975). Figure 11.2 shows a molecule of chloroplast DNA from Spinacia oleracea. The significance of circularity in DNA is not known, but it is a useful property since it implies the molecule has not been degraded on isolation. The most interesting aspect of this finding is that in all the species listed the contour length of the majority of the circles is in the range 37–45 µm; this length of double-stranded DNA has a calculated molecular weight of 0.85–1.0 × 108 daltons, which is in the same range as the kinetic complexity of chloroplast DNA (Table 11.2). This correspondence of length and kinetic complexity suggests that the genetic information carried by the chloroplast DNA is accommodated by the length of the circular molecule.

Figure 11.2
Open circular DNA molecule, contour length
44.7 µm, from chloroplasts of Spinacia oleracea.
(Reprinted from Hermann et al., (1975) by courtesy of Elsevier.)
11.2.1.6—
Ploidy
It is clear from the data in Table 11.2, and from the evidence for circular DNA, that each chloroplast may contain many copies of a circular genome, and thus may be genetically polyploid. It has been pointed out by Kirk (1972) that this
multiplicity increases the probability that a mutation will appear in a chloroplast in a given time, but there is likely to be a long delay before the mutation can be expressed, since a mutation in one copy will be swamped by all the other wild-type copies. It is therefore not surprising that known non-Mendelian mutations affecting chloroplasts are small in number, and difficult to induce with mutagens. In Chlamydomonas reinhardi a number of mutations are known which alter the sensitivity of chloroplast ribosomes to inhibition by antibiotics such as erythromycin and carbomycin; some of these mutations show uniparental inheritance (Sager, 1972; Mets & Bogorad, 1972). Those genes which are inherited in a uniparental fashion have been shown by recombinational analysis to form one linkage group which behaves as if it were diploid in vegetative cells. It is difficult to reconcile this diploid behaviour with the evidence which suggests that there are many copies of the genome in each chloroplast. It may be that the uniparental linkage group does not reside in chloroplast DNA, or if it does, that a reduction in the number of genomes in each chloroplast occurs at some stage in the cell cycle, so that only a 'master copy' is passed during sexual reproduction. There is as yet no evidence to resolve these questions.
11.2.1.7—
Functions
A 40 µm circle of double-stranded DNA of unique base sequence is sufficient in principle to code for about 125 proteins each of molecular weight 50,000. Since our knowledge depends ultimately on the validity of the techniques which can be used, the evidence for the functions of chloroplast DNA will be considered in terms of the four methods tried so far:
The Selective Inhibition of Chloroplast DNA Transcription
The antibiotic rifampicin is a potent inhibitor of the initiation of RNA synthesis in bacteria. This compound has been reported to inhibit the incorporation of labelled precursors into chloroplast ribosomal-RNA, but not into cytoplasmic ribosomal-RNA, in several unicellular algae, namely Chlamydomonas, Chlorella, and Acetabularia. Rifampicin has also been reported to inhibit chloroplast RNA polymerase activity in extracts of Chlamydomonas. These results suggest that the functional genes for chloroplast ribosomal-RNA are located in chloroplast DNA and are transcribed by the chloroplast RNA polymerase.
The effect of rifampicin in higher plant systems is controversial. There have been some reports that it specifically inhibits chloroplast ribosomal-RNA synthesis, but other workers could not reproduce this result (Bottomley et al., 1971). There now seems general agreement that chloroplast RNA polymerase from higher plants, as normally prepared and assayed, is insensitive to rifampicin, but this may mean only that the preparations are not initiating RNA synthesis.
Genetic Analysis of Mutants
Many mutations are known which affect chloroplast components, but the vast majority are inherited in a Mendelian fashion and are therefore presumed, to be located in nuclear genes. For example, seven different genes are known which control steps in the chlorophyll biosynthetic pathway, and nuclear genes have been shown to be involved in the synthesis of phosphoribulokinase and at least five components of the photosynthetic electron transport chain (Kirk, 1972; Kirk & Tilney-Bassett, 1967; Levine & Goodenough, 1970).
In Chlamydomonas many mutations affecting chloroplast ribosomes and photosynthetic capacity are inherited as a single linkage group in a uniparental fashion (Sager, 1972; Schlanger & Sager, 1974). In this alga, like gametes fuse completely, and the physical basis for uniparental inheritance has been suggested to be the destruction of the chloroplast DNA of one of the gametes by a modification-restriction system of the type known in bacteria. As pointed out earlier, there are difficulties in concluding that this uniparental class of mutations resides in chloroplast DNA, but it is tempting to believe that it does. A simple interpretation suggests that some of the proteins of the chloroplast ribosomes are encoded in chloroplast DNA; there is also evidence that at least one other protein of the chloroplast ribosome is encoded in a nuclear gene (Mets & Bogorad, 1972).
Rigorous proof that a mutation inherited in a maternal or uniparental fashion actually alters the amino acid sequence of a protein has been obtained in only one case. Chan and Wildman (1972) studied the inheritance of a mutation in the large subunit of Fraction I protein in Nicotiana tabacum at the tryptic peptide level; this mutation was inherited via the maternal line only. By contrast, mutations in the small subunit of Fraction I protein are inherited in a Mendelian fashion (Kawashima & Wildman, 1972). This type of combined biochemical-genetic approach is very promising and deserves further use, especially with cultivated plants where many varieties are available. It must be emphasized that this approach must be conducted at the level of the tryptic peptide analysis of a purified protein. It is not sufficient to show that particular proteins are absent in a mutant variety, since absence of a protein might result from a mutation in a chloroplast gene which controls the formation of a protein encoded in the nucleus.
DNA-RNA Hybridization Studies
If RNA isolated from chloroplasts can be shown to hybridize to chloroplast DNA but not to nuclear DNA, this is good grounds for believing that such RNA is both encoded in and transcribed from chloroplast DNA. A number of hybridization studies have been carried out with chloroplast ribosomal-RNA, which hybridizes to 0.5–6.0% of chloroplast DNA (Ellis & Hartley, 1974). The
most recent study is that of Thomas and Tewari (1974); they found that in a number of higher plants each circle of chloroplast DNA contains two genes for chloroplast ribosomal-RNA. There are several reports that chloroplast ribosomal-RNA will also hybridize to nuclear DNA. The significance of these reports awaits further study, but the possibility is raised that there are two types of chloroplast ribosomal-RNA, one encoded in chloroplast DNA and the other in nuclear DNA. This arrangement could be a means whereby the nucleus exerts control over events in the chloroplasts, especially if it is further postulated that chloroplast ribosomes containing nuclear RNA translate only messengers originating in the nucleus.
There is evidence that some of the transfer RNA species found in chloroplasts will also hybridize to chloroplast DNA. Tewari and Wildman (1970) found that tRNA from tobacco chloroplasts would hybridize to 0.4–0.7% of chloroplast DNA. This amount of DNA would be enough to code for 20 to 30 transfer RNA molecules, each of molecular weight 25,000. This work used unfractionated tRNA however, and more studies need to be done to establish the site of encoding of individual tRNA species.
Identification of RNA and Protein Molecules Synthesized by Isolated Chloroplasts
This is the most direct method; if transcription coupled to translation can be obtained in isolated chloroplasts, identification of the products would simultaneously determine both the structural genes present in chloroplast DNA and the function of chloroplast ribosomes. Work in the author's laboratory has shown that isolated intact chloroplasts from Pisum sativum and Spinacia oleracea will synthesize discrete protein and RNA molecules, but transcription and translation are not coupled (Hartley & Ellis, 1973; Ellis, 1974; Ellis & Hartley, 1974). It is therefore not possible to infer that the proteins synthesized by isolated chloroplasts are encoded in chloroplast DNA. The RNA synthesized by isolated chloroplasts has been analyzed by polyacrylamide gel electrophoresis (Fig. 11.3). The chief product is a species of molecular weight about 2.7 × 106 ; this has been shown by competitive hybridization to chloroplast DNA to be a precursor to chloroplast ribosomal-RNA. Isolated chloroplasts have not been shown convincingly to synthesize ribosomal-RNA; presumably the processing system which trims the precursor molecules does not survive the trauma of isolation (see also chapter 9).
The known genes in chloroplast DNA can be summarized as follows: chloroplast transfer-RNA, chloroplast ribosomal-RNA, several chloroplast ribosomal proteins, and the large subunit of Fraction I protein. These genes account for less than 10% of the total potential coding capacity of a 40 µm circle of DNA. The elucidation of the function of the remaining 90% of chloroplast DNA is the most important problem in this field.
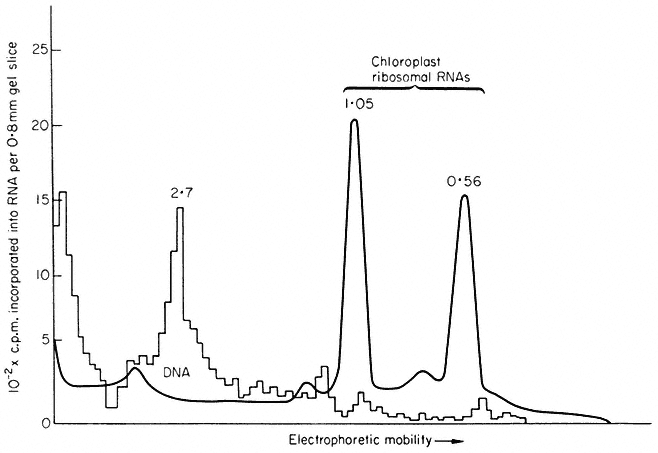
Figure 11.3
Analysis by polyacrylamide gel electrophoresis of RNA synthesized by
chloroplasts isolated from Spinacia oleracea. Isolated chloroplasts were incubated
with 3 H-uridine and 15,000 lux of red light for 20 min at 20°C. Nucleic acid was extracted
and run on polyacrylamide gels. The solid line represents the A260 , while the histogram
shows the radioactivity. The figures are the molecular weights × 10-6 of the RNA components.
(Reprinted from Hartley & Ellis, 1973, by courtesy of the Biochemical Journal).
11.2.2—
Chloroplast DNA Polymerase
DNA polymerase activity has been detected in chloroplast suspensions prepared from Spinacia oleracea. Nicotiana tabacum, and Euglena gracilis. The product renatures readily after heat denaturation, and hybridizes to a much larger extent with chloroplast DNA than with nuclear DNA. It has not been established whether the chloroplast polymerase is identical with any nuclear polymerase or whether it carries out either a replicase or a repair function. The enzyme is bound to the chloroplast membranes, but in the case of Euglena, it has been solubilized by treatment with high concentrations of salt and highly purified; the purified enzyme is inhibited by ethidium bromide (Keller et al., 1973). Flechtner and Sager (1973) have made the interesting observation that treatment of Chlamydomonas cells with ethidium bromide induces a selective and reversible inhibition of chloroplast DNA replication. Thus nuclear DNA synthesis proceeds normally while replication of chloroplast DNA is impaired; the preexisting chloroplast DNA decreases by at least 80% in one cell generation, but this loss is reversible if the drug is removed within 12 hours. This result suggests that one or a few of the chloroplast DNA copies may be protected in some way,
perhaps by close attachment to chloroplast membranes. Such sequestered DNA could act as a 'master' copy, and might account for the diploid behaviour of the uniparental linkage group in this organism. The chloroplast DNA of Euglena and Chlamydomonas has been shown to replicate in a semi-conservative fashion at a different time in the growth cycle from nuclear DNA, but the factors controlling the time and rate of synthesis of chloroplast DNA are unknown.
11.2.3—
Chloroplast RNA Polymerase
DNA-dependent RNA polymerase activity has been demonstrated in chloroplast preparations from several species of algae and higher plants. Most workers study lysed chloroplast preparations because the nucleotide triphosphates used as substrates penetrate the chloroplast envelope at slow rates. Unlike intact
Figure 11.4
Distribution of chloroplast RNA polymerase activity in leaves of
young shoots of Pisum sativum. Pea seedlings were grown for 14 days
and the leaves homogenized. Total RNA polymerase activity was measured
in the low-speed pellet containing nuclei and chloroplasts, and chloroplast
RNA polymerase activity in purified chloroplast membranes. The figures are the
activities of the chloroplast RNA polymerase as percentages of the total activity.
(Unpublished work of J. Bennett.)
chloroplasts, which use light to phosphorylate added 3 H-uridine (Fig. 11.3), lysed preparations do not synthesize discrete species of RNA; instead a polydisperse pattern of products ranging in size from 5s to 23s is obtained (Spencer & Whitfeld, 1967). This difference between the products from intact and lysed chloroplasts may result from the dilution of controlling factors on lysis.
The RNA polymerase activity of chloroplast preparations can represent a high proportion of the total RNA polymerase activity measurable in leaf extracts. Figure 11.4 illustrates this point in the case of the growing shoot of Pisum sativum; the chloroplast polymerase activity can account for half of the total activity in the youngest leaves at the stem apex, but accounts for progressively less in the older leaves.
There are two key questions about chloroplast RNA polymerase which need to be answered: firstly, how similar is it to any nuclear polymerase, and secondly, how is its activity regulated ? The enzyme is bound to the chloroplast lamellae, but a technique for its quantitative removal has been devised (Bennett & Ellis, 1973). The solubilized chloroplast polymerase has been purified, and appears very similar to a nuclear polymerase (Bogorad et al., 1973), but a detailed characterization has yet to be reported. It has been shown that the synthesis of chloroplast ribosomal-RNA in dark-grown plants is phytochrome-mediated (Scott et al., 1971), but the nature of the mechanism which links phytochrome to RNA polymerase is unknown.
11.2.4—
Chloroplast Protein Synthesis
Chloroplast preparations capable of incorporating labelled amino acids into protein have been isolated from higher plants and algae. Table 11.3 illustrates some results obtained with preparations from young bean and tobacco leaves; preparations from mature leaves have little activity. These results are typical of those reported in many studies with many species (Ellis et al., 1973). In such work it is vital to establish that the incorporation is due to chloroplasts and not to microorganisms, intact leaf cells, or cytoplasmic ribosomes. The best criterion is the dependence of incorporation on an added energy source; this is ATP in the case of lysed chloroplasts, or light in the case of intact chloroplasts which can carry out photophosphorylation. Dependence on an added energy source eliminates intact leaf cells and microorganisms as the agents of incorporation. Activity by contaminating cytoplasmic ribosomes can be ruled out by the different sensitivity of the two types of ribosome to antibiotic inhibitors (see 11.2.4.5). Other criteria are the sensitivity of incorporation to added ribonuclease, and to variation in the concentration of added Mg2+ ions; these criteria are useful when lysed chloroplasts or isolated ribosomes are being tested, but not when the incorporation is due to intact chloroplasts.
Some of the characteristics and functions of protein synthesis by chloroplasts will now be considered.
| ||||||||||||||||||||||||||||||
11.2.4.1—
Ribosomes
Lyttleton (1962) was the first to show that green plant cells contain two classes of ribosome which differ in their sedimentation coefficients. Chloroplasts contain 70s ribosomes while the cytoplasm contains 80s ribosomes. Figure 11.5 shows a density gradient analysis of an homogenate of spinach leaves, from which it can be seen that chloroplast ribosomes constitute a high proportion of the total cellular complement of ribosomes; values as high as 60% have been reported.
Chloroplast ribosomes resemble those from prokaryote cells in their s value. Their RNA components are also of the same size as those found in prokaryote ribosomes, i.e. 16s and 23s. The 80s cytoplasmic ribosomes by contrast, contain 18s and 25s RNA molecules. These similarities between chloroplast and prokaryote ribosomes have been much stressed in the past, but it is now clear that these similarities are of size only, and not in the primary structure of the RNA and protein components. For example, ribosomal-RNA from Escherichia coli does not compete with chloroplast ribosomal-RNA from Euglena for hybridization to chloroplast DNA. The protein complement of prokaryote ribosomes is also quite different from that of chloroplast ribosomes as judged by gel electrophoresis and immunological tests. Similarly, the proteins of chloroplast and cytoplasmic ribosomes from the same plant differ distinctly in the patterns they give on gel electrophoresis.
Some of the chloroplast ribosomes are bound to the internal membranes. Electron microscopy has revealed whorl-like polyribosomes attached to the granal and intergranal thylakoid membranes, while analysis of isolated chloroplasts shows that up to 50% of the ribosomes cannot be removed from the membranes by washing with hypotonic buffer. Tao and Jagendorf (1973) have
Figure 11.5
Proportion of chloroplast ribosomes in leaves of Spinacia
oleracea. Extracts of spinach leaves were treated with
detergent to release membrane-bound ribosomes, and all the
ribosomes collected by centrifugation. The ribosomes
were analysed on a linear sucrose density gradient.
(Unpublished work of M. R. Hartley.)
shown that some free ribosomes are lost during chloroplast isolation without the chloroplasts lysing irreversibly; when this loss is taken into account, they estimate that in chloroplasts from Pisum sativum, about 20% of the ribosomes are membrane-bound while the remainder are free in the stroma. There is evidence that the two classes of chloroplast ribosome synthesize different types of protein (see 11.2.4.6).
11.2.4.2—
Amino Acid Activation
Chloroplasts isolated from several algae and higher plants have been found to contain aminoacyl-transfer-RNAs and the corresponding synthetases (Ellis et al., 1973). These transfer RNA and synthetase molecules can often, but not always, be distinguished from those involved in the activation of the same amino acids in the cytoplasm. Studies of the compatibility between transfer-RNA and synthetases from the two cellular compartments show that the specificity of interaction ranges from. none to absolute. For example, the cytoplasm of leaves of Phaseolus vulgaris contains a leucyl-transfer-RNA synthetase which can add leucine only to the two leucyl-transfer-RNA species common to the cytoplasm and chloroplasts, but is not able to recognize the three leucyl-transfer-RNA
species found only in the chloroplasts. The chloroplast enzyme, on the other hand, can aminoacylate all five leucyl-transfer-RNA species. It is difficult to attach any biological significance to these variations in specificity. The simplest presumption is that in the intact cell the chloroplast synthetases aminoacylate only the transfer-RNA species located in the chloroplast and these are sufficient for chloroplast protein synthesis to proceed, while the cytoplasmic synthetases aminoacylate only the transfer-RNA species located in the cytoplasm. A more interesting possibility is that some of the transfer RNA species required to translate chloroplast messenger-RNA are acylated in the cytoplasm; such a requirement would provide one way in which protein synthesis in the cytoplasm and chloroplast could be integrated, but there is no direct evidence to support this suggestion. Experiments with Euglena suggest that at least some of the chloroplast aminoacyl-transfer-RNA synthetases are encoded in nuclear DNA and are synthesized on cytoplasmic ribosomes (Hecker et al., 1974).
11.2.4.3—
Initiation of Chloroplast Protein Synthesis
Protein synthesis in bacteria is initiated by N-formyl-methionyl transfer RNA; there is evidence that this is also true in chloroplasts, but not in the cytoplasm of eukaryotic cells. Schwarz et al., (1967) found that chloroplast ribosomes from Euglena would translate RNA from bacteriophage f2 into viral coat protein with N-formylmethionine at the amino terminus. Detailed studies of initiating methionyl-transfer-RNA species have since been reported for several higher plants. For example, there are two methionyl-transfer-RNA species in wheat chloroplasts. One of these can be formylated by a chloroplast transformylase, whereas the other cannot, and may serve to direct methionine into internal positions in the polypeptide chains (Leis & Keller, 1971). Two other methionyl-transfer-RNA species are found in the cytoplasmic fraction, neither of which can be formylated. It is probable from this type of evidence that the initiation of protein synthesis by chloroplasts is similar to that in prokaryotes in that it uses a formylated methionyl-transfer-RNA, but distinct from that in the cytoplasm which uses an unformylated methionyl-transfer-RNA. The significance of this distinction is not clear; it may reflect the evolutionary origin of the compartments in eukaryotic cells, and have no contemporary functional value.
11.2.4.4—
Energy Source
In most studies of protein synthesis by isolated chloroplasts, added ATP has been used as an energy source (see Table 11.3). Since isolated chloroplasts can generate ATP by photophosphorylation, it should be possible to drive protein synthesis in chloroplasts with light. Spencer (1965) found that spinach chloroplasts would use light to incorporate amino acids into protein provided that ADP, inorganic phosphate and pyocyanine were added. The necessity for
pyocyanine as catalyst indicates that the chloroplasts were broken, and had lost their natural catalyst, ferredoxin, by dilution. If precautions are taken to isolate chloroplasts which have their outer envelopes intact, protein synthesis is stimulated twenty-fold by light in the absence of either cofactors or catalysts of photophosphorylation (Fig. 11.6). The rates of light-driven protein synthesis
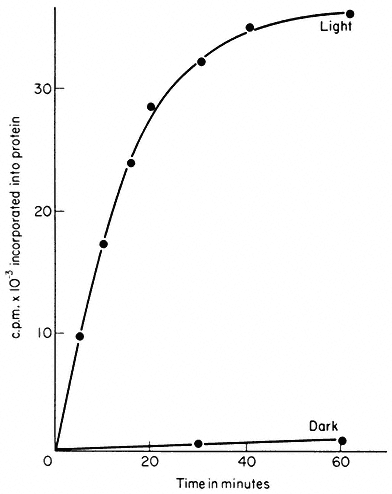
Figure 11.6
Light-driven protein synthesis by chloroplasts isolated from Pisum sativum.
Intact isolated chloroplasts were incubated with 14 C-leucine and 4,000 lux red
light at 20°C. Total protein was then extracted and its radioactivity measured.
(Reprinted from Blair & Ellis, 1973, by courtesy of Elsevier.)
by such chloroplasts are the highest yet recorded for any isolated chloroplast system. The use of intact chloroplasts for studies of protein synthesis has a major advantage over the use of broken chloroplasts in that conditions around the polysomes are more normal with respect to controlling factors. It is thus more likely that correct termination and release of polypeptide chains occurs in intact chloroplasts than in lysed preparations; such intact chloroplasts make discrete protein molecules rather than incomplete polypeptide chains (see 11.2.4.6). A further advantage of using light as the energy source is that the broken chloroplasts, which inevitably contaminate the preparation of intact chloroplasts, cannot contribute to the incorporation in the absence of added cofactors.
It is possible that protein synthesis by chloroplasts in vivo uses ATP provided by the chloroplast itself. However, this is not the case in developing
chloroplasts, since organisms such as Chlorella, Chlamydomonas, and Pinus do not require light to form chloroplasts. The formation of chloroplasts must therefore depend on ATP supplied by the rest of the cell. This conclusion is confirmed by the ability of intact isolated etioplasts from Pisum sativum to use ATP, but not light, as their source of energy for protein synthesis (Siddell & Ellis, 1975).
11.2.4.5—
Inhibitors
It is well established that protein synthesis by isolated chloroplast ribosomes is inhibited by the same antibiotics which inhibit protein synthesis by prokaryote ribosomes. The best studied example is chloramphenicol, which inhibits protein synthesis by isolated chloroplasts from all the species so far tested; inhibition is shown only by the D-threo isomer of chloramphenicol, and not by the other stereo isomers. Three other unrelated antibiotics, spectinomycin, lincomycin, and erythromycin also inhibit protein synthesis by chloroplast ribosomes. Light-driven protein synthesis by chloroplasts from Pisum sativum is especially sensitive to lincomycin, 50% inhibition being given by 0.2 µg ml–1 (Ellis & Hartley, 1971). Protein synthesis by cytoplasmic ribosomes from plants is not inhibited by any of these antibiotics. On the other hand, cytoplasmic ribosomes are inhibited by cycloheximide, which does not affect the activity of chloroplast ribosomes. It must be emphasized that all these findings relate only to the activity of isolated sub-cellular systems; it cannot be assumed from these data alone that it is valid to use these compounds on intact cells with the expectation of inhibiting protein synthesis in one compartment but not in the other.
The protein complements of chloroplast and bacterial ribosomes are different, and thus the above similarity between them must reside in their antibioticbinding sites, and not in the properties of the proteins as revealed by gel electrophoresis or immunological tests. There is no evidence that the mechanism of action of bacterial antibiotics on chloroplast ribosomes is similar to that on bacterial ribosomes; nor is there any understanding of the selective pressure which maintains the sensitivity of chloroplast ribosomes to these antibiotics in the face of mutations to resistance, which can, for example, be found in Chlamydomonas (see 11.2.1.6).
11.2.4.6—
Functions
The high proportion of plant ribosomes located inside chloroplasts raises the question as to their function. Are they required in such quantities because they make a large number of different chloroplast proteins, or because they make a small number of different chloroplast proteins in large amounts? The available evidence suggests that the latter is the case (Ellis et al., 1973; Ellis, 1974).
The problem of identifying which proteins are synthesized by chloroplast ribosomes has been tackled in two ways. The first is to supply antibiotic inhibitors, especially chloramphenicol and cycloheximide, to intact cells making chloroplasts, and determining which proteins are no longer synthesized. The validity of results from experiments with any inhibitor depends absolutely on the specificity of its action in intact cells, and there is evidence that both chloramphenicol and cycloheximide have effects on activities other than protein synthesis in some higher plants (Ellis & MacDonald, 1970). Processes such as ion uptake, oxidative phosphorylation and photophosphorylation are inhibited by all four stereoisomers of chloramphenicol, whereas the inhibition of protein synthesis by isolated chloroplast ribosomes is specific for the D -threo isomer (Ellis, 1969). This stereospecificity provides a means of establishing for any particular tissue whether chloramphenicol is inhibiting protein synthesis directly at the ribosomal level, or in addition, is affecting some other process such as energy supply. It is strongly recommended that only if an inhibition is produced specifically by the D -threo isomer should an interpretation directly involving protein synthesis be invoked. Another problem is that in most of the inhibitor experiments on chloroplast ribosomal function, increases in specific proteins have been measured as enzymic activities rather than as amounts of protein. Failure to observe an effect by a particular inhibitor might therefore mean that the increase in enzymic activity is due to activation of a precursor protein, rather than to de novo synthesis by either chloroplast or cytoplasmic ribosomes.
Bearing these difficulties in mind, this author interprets the bulk of the published inhibitor experiments as suggesting that most of the chloroplast proteins are synthesized, on cytoplasmic ribosomes (Ellis et al., 1973; Ellis, 1975). In all the studies reported on several algae and higher plants, the synthesis of Fraction I protein was found to be inhibited by 70s ribosomal inhibitors. In most cases, the synthesis of the other soluble enzymes of the photosynthetic carbon dioxide reduction cycle appears to occur on cytoplasmic ribosomes; the same is true for ferredoxin and the chloroplast RNA polymerase. Besides Fraction I protein, the only other proteins which appear to be synthesized by chloroplast ribosomes are some of the chloroplast ribosomal and lamellar proteins, including the photosynthetic cytochromes. It must be emphasized, however, that these inhibitor experiments are never more than suggestive. Strictly interpreted, they never say more than that the activity of a particular group of ribosomes is required for a particular protein to accumulate in the chloroplast. This is not the same as saying that these ribosomes actually synthesize that protein, because it is possible that the apoenzyme is synthesized by one class of ribosomes but requires for its appearance in the chloroplast in an active state, additional protein(s) which are synthesized by another class of ribosomes.
That this is true for Fraction I protein has been shown by the second approach to the problem of chloroplast ribosomal function, namely, the
synthesis of specific proteins by isolated sub-cellular systems. This is the most direct approach to this problem but it has been successful only recently because of the difficulty of isolating sub-cellular systems which will carry out complete protein synthesis with fidelity; algal systems are especially difficult in this respect. Isolated intact chloroplasts from Pisum sativum, however, synthesize at least six discrete proteins (Fig. 11.7); similar results have been obtained with
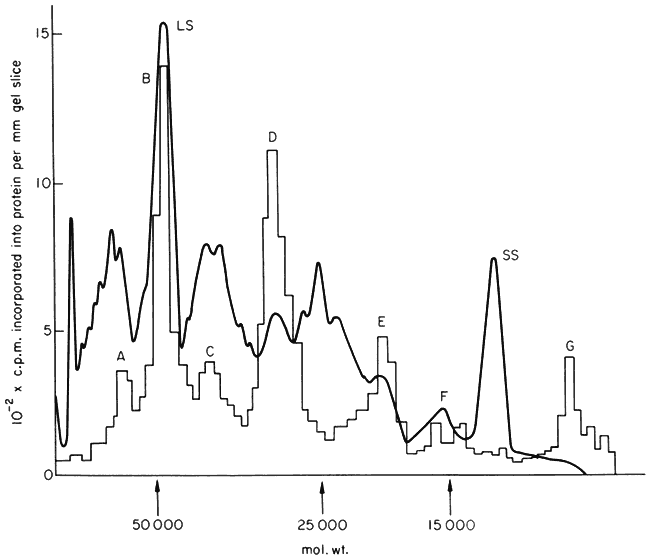
Figure 11.7
Products of protein synthesis by chloroplasts isolated from Pisum sativum.
Chloroplasts were incubated as in Fig. 11.6, and then dissolved in sodium
dodecylsulphate. The solubilized extract was electrophoresed on gels containing
15% polyacrylamide. The smooth line represents the absorbance of stained protein
bands, and the histogram is radioactivity. The letters A–G represent the discrete
radioactive peaks. LS and SS mark the large and small subunit of Fraction I
protein respectively. (From Eaglesham & Ellis, 1974, by courtesy of Elsevier.)
chloroplasts from Hordeum vulgare and Spinacia oleracea. Only one of these proteins (peak B in Fig. 11.7) is soluble; it has been identified as the large subunit of Fraction I protein by tryptic peptide analysis (Blair & Ellis, 1973). Recent work has shown this large subunit to be made by the free ribosomes of the chloroplast, but not by the membrane-bound ribosomes. The high proportion of chloroplast ribosomes found in leaves may thus be required because Fraction I protein is one of the most abundant proteins (Ellis, 1973). The small
subunit of Fraction I protein is not synthesized by isolated chloroplasts, but it has been detected as the product of protein synthesis by isolated cytoplasmic ribosomes from Phaseolus vulgaris leaves (Kekwick & Gray, 1973). The other proteins made by isolated chloroplasts are all membrane-bound, and have so far resisted identification; it is clear they are minor components of the thylakoids. Two proteins associated with the chloroplast envelope are also labelled in isolated pea chloroplasts (Joy & Ellis, 1975). The same pattern of proteins synthesized by intact pea chloroplasts has been found in studies of protein synthesis by isolated pea etioplasts (Siddell & Ellis, 1975).
This in vitro approach to the study of chloroplast ribosomal function gives direct and unambiguous results, and it should be extended to species lower on the evolutionary scale to see how far the pattern found in angiosperms extends. In view of the uniformity of the size of the chloroplast genome in algae and higher plants (Table 11.2), it seems likely that the pattern seen in isolated pea chloroplasts will be universal. However, this assumes that proteins synthesized by chloroplast ribosomes are also encoded by chloroplast DNA; this may not be true if messenger-RNA can cross the chloroplast envelope, but so far this is only a theoretical possibility.
Another area which may repay attention is the study of protein synthesis by plastids other than chloroplasts, especially proplastids and amyloplasts. It seems probable that these plastids synthesize a different range of proteins from chloroplasts. If these proteins are encoded in plastid DNA, they would account for some of the functions of the 90% of this genome which have not been identified (see 11.2.1.7).
11.2.5—
Co-Operation between Chloroplast and Nuclear Genomes
It is clear from the available evidence that the chloroplast genome requires for its expression the co-operation of the nuclear genome. The best information about the details of this co-operation concerns the synthesis of Fraction I protein. By combining the results of Wildman's genetic studies with those of the in vitro studies of protein synthesis, a model for the synthesis of Fraction I protein can be constructed (Fig. 11.8). In this model, the large subunit is both encoded and synthesized within the chloroplast, while the small subunit is both encoded and synthesized outside the chloroplast. This model is tidy, and requires protein, but not nucleic acid, to cross the chloroplast envelope. This transport of protein must be on a large scale because it involves not only the small subunit of Fraction I protein but all the other proteins which, from the inhibitor evidence, are made on cytoplasmic ribosomes. The mechanism of this transport is unknown, but it must be able to distinguish between different proteins. One suggestion is that a protein exists in the outer envelope of the chloroplast which recognizes a site common to all those proteins made on cytoplasmic ribosomes, but destined to function in the chloroplast.
Besides structural and enzymic proteins, regulatory proteins probably also cross the chloroplast envelope. How light regulates chloroplast development is not known, but it would be reasonable to speculate in terms of proteins entering the organelle to trigger nucleic acid and protein synthesis. There is the possibility of proteins also passing out of the organelle, since recent work in Chlamydomonas has implicated chloroplast protein synthesis in the regulation of nuclear DNA replication (Blamire et al., 1974). This question of protein transport into and out of the chloroplast is a crucial one to study if we are to understand how the chloroplast and nuclear genomes interact; almost no research has been carried out in this area. The movement of nucleic acids across the envelope remains a possibility, but no compelling evidence to support it has been published.
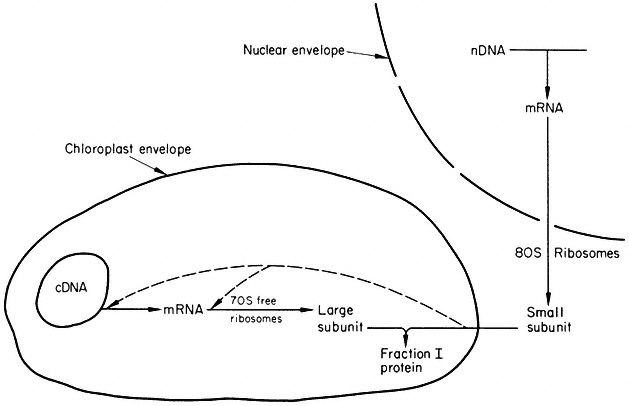
Figure 11.8
Model for the co-operation of nuclear and chloroplast genomes in the
synthesis of Fraction I protein. cDNA and nDNA stand for chloroplast and
nuclear DNA respectively. The dashed lines indicate possible sites at
which small subunits may control the synthesis of large subunits.
(Reprinted from Ellis, 1975, by courtesy of Pergamon Press.)
Another problem needing study is the nature of the mechanism which integrates the synthesis of the two subunits of Fraction I protein in different cellular compartments. The dashed lines in Fig. 11.8 illustrate one possibility; the small subunit is postulated to be required as a positive factor for the initiation of either transcription or translation of the large subunit messenger (Ellis, 1975). The recent detection of the large subunit messenger should allow this possibility to be tested (Hartley et al., 1975).
11.3—
Mitochondrial Autonomy
Research on the biogenesis of plant mitochondria has lagged far behind that on mitochondria from animal and fungal cells. A summary of the position is given here in the hope of stimulating readers to undertake work in this field; recent reviews have appeared on the biogenesis of both animal and fungal mitochondria (Borst, 1971, 1972), and of those in plants (Boulter et al., 1972; Leaver & Harmey, 1973; Leaver, 1975).
The first point to emphasize is that mitochondria constitute a much smaller compartment in plant cells compared to chloroplasts. The amount of mitochondrial DNA and ribosomes is correspondingly much smaller as a percentage of the total cellular complement. The technical difficulties of extracting small quantities of intact, uncontaminated mitochondria from plant tissues make studies on their genetic system more difficult than in the case of chloroplasts. Another difference from chloroplasts is that the DNA and ribosomes of mitochondria are more variable in their properties between species, so the extrapolation of results must be done with caution.
In a range of higher plants the density of mitochondrial DNA is constant at 1.706–1.707 g cm–3 , whereas the nuclear DNA from the same species varies in density between 1.691–1.702 g cm––3 . In many organisms mitochondrial DNA has been isolated in circular form; in contrast to the situation for chloroplasts (see 11.2.1.5), the contour length of the circles varies with the species. Thus, animals have mitochondrial DNA circles of about 5 µm contour length; this corresponds to a molecular weight of about 107 , or 15,000 base pairs. In Ascomycetes the contour length of circular mitochondrial DNA is in the range 20–26 µm, while in higher plants lengths of 30 µm have been reported. The small size of the mitochondrial DNA in animals means that it can code for only a few components. Hybridization studies in Xenopus indicate that mitochondrial ribosomal RNA and at least some mitochondrial transfer RNA are encoded in mitochondrial DNA. The remaining mitochondrial DNA is sufficient to code for only about ten proteins each of molecular weight 50,000. However, kinetic complexity measurements on mitochondrial DNA from higher plants indicate that it has between six and ten times the coding potential of animal mitochondrial DNA. This observation raises the possibility that mitochondria from plants possess a higher degree of autonomy than those from animals, and this reinforces the plea that more research be carried out with plant cells.
The sedimentation coefficients of mitochondrial ribosomes and their component ribosomal-RNAs vary greatly with the species. Animal mitochondria contain so-called 'miniribosomes' which sediment between 55s and 60s, while fungal mitochondria contain 70–74s ribosomes. Leaver and Harmey (1972) have shown that mitochondrial ribosomes from several higher plants sediment at 77s–78s, while their major RNA components sediment at 25s and 18s. Although mitochondrial ribosomes differ in size in different species, they are all similar
in that protein synthesis by them is sensitive to the same set of antibiotics which inhibit prokaryote and chloroplast ribosomes.
Protein synthesis by isolated mitochondria has been studied extensively in animals and fungi, but hardly at all in higher plants. Several proteins of the inner, but not the outer, mitochondrial membrane are synthesized by animal and fungal mitochondria. These proteins have been identified as subunits of the ATPase complex, cytochrome b, and cytochrome c oxidase; the other subunits of these insoluble complexes are synthesized by cytoplasmic ribosomes. This evidence suggests the synthesis of mitochondrial membranes requires the co-operative activity of both mitochondrial and nuclear genomes. This idea of the co-operation between the several genetic systems in eukaryotic cells is already familiar to the reader from section 11.2.5. The same problem of the transport of proteins across the bounding membranes applies to mitochondria as well as to chloroplasts. In contrast to chloroplasts, mitochondria from animal and fungal cells synthesize no soluble proteins; whether higher plant mitochondria synthesize any soluble proteins is unknown.
No protein has so far been rigorously identified as being encoded in mitochondrial DNA in any organism. A simple presumption is that those proteins synthesized by isolated mitochondria are encoded in mitochondrial DNA. It is likely that the integrative controls between nuclear and mitochondrial genomes in higher plants differ from those in animals which contain less than one sixth as much DNA. Why do plant mitochondria apparently contain so much more genetic information to perform the same functions as animal mitochondria? Answering this question is a challenge which must be taken up if our understanding of the origin of this organelle in plants is to improve.
Further Reading
Borst P. (1971) Size, structure and information content of mitochondrial DNA. In Autonomy and Biogenesis of Mitochondria and Chloroplasts, (eds N.K. Boardman, A.W. Linnane & R.M. Smillie), pp. 260–6. North Holland, Amsterdam.
Borst P. (1972) Mitochondrial nucleic acids. Ann. Rev. Biochem.41, 333–76.
Boulter D., Ellis R.J. & Yarwood A. (1972) Biochemistry of protein synthesis in plants. Biol. Rev.47, 113–75.
Ellis R.J. (1975a) The synthesis of chloroplast membranes in Pisum sativum. In Membrane Biogenesis: Mitochondria, Chloroplasts & Bacteria, (ed. A. Tzagoloff). Plenum Publishing Co., New York.
Ellis R.J., Blair G.E. & Hartley M.R. (1973) The nature and function of chloroplast protein synthesis. Biochem. Soc. Symp.38, 137–62.
Ellis R.J. & Hartley M.R. (1974) Nucleic acids of chloroplasts. In Biochemistry of Nucleic Acids, (ed. K. Burton). MTP International Review of Science Biochemistry Series One, Vol. 6, 323–48. Butterworths, London & University Park Press, Baltimore.
Kirk J.T.O. (1972) The genetic control of plastid formation: recent advances and strategies for the future. Sub-Cell. Biochem.1, 333–61.
Leaver C.J. (1975) The biogenesis of plant mitochondria. In The Chemistry and Biochemistry of Plant Proteins, (ed. J. Harborne). pp. 137–65. Academic Press, New York & London.
Leaver C.J. & Harmey M.A. (1973) Plant mitochondrial nucleic acids. Biochem. Soc. Symp.38, 175–93.
Levine R.F. & Goodenough U.W. (1970) The genetics of photosynthesis and of the chloroplast in Chlamydomonas reinhardi. Ann. Rev. Genet,4, 397–408.
Sager R. (1972) Cytoplasmic Genes and Organelles. Academic Press, New York & London.
Tewari K.K. & Wildman S.G. (1970) Information content in the chloroplast DNA. In Control of Organelle Development, (ed. P.L. Miller), pp. 147–79. Symposium 24 of the Society for Experimental Biology, Cambridge University Press, Cambridge.
Chapter 12—
Regulation of Enzyme Levels and Activity
12.1—
Introduction
The first theory of metabolic control was introduced into plant physiology in 1905 by F. F. Blackman and has become known as the 'Law of Limiting Factors' which states that 'when a process is conditioned as to its rapidity by a number of separate factors the rate of the process is limited by the pace of the slowest reaction'.
It is not possible to derive a rate equation from this statement and discussions of the 'Law' tend to be by analogy, e.g. the strength of a chain is the strength of its weakest link. Experimentally, the statement has led to the fruitless search for metabolic master reactions. Mathematically, we can derive rate equations for multistep reactions and establish that every step contributes to the overall rate. Theoretically Blackman's statement is a denial of the steady state; if a metabolic sequence is in a steady state then the concentration of each intermediate is constant and the individual reactions proceed at the same rate—hence no reaction can be described as the slowest.
12.1.1—
Pacemakers
If in a metabolic sequence all reactions are proceeding at the same pace then a single reaction may be almost entirely responsible for determining that pace; such a reaction has been called a 'bottleneck' or pacemaker and much work in metabolic control is directed towards the identification of such control points. Krebs and Kornberg (1957) in a consideration of pacemakers stated 'there is a principle which may guide the search for pacemakers. As pacemakers are reactions of variable rate, the level of substrate concentration of the pacemaker must vary inversely with the rate: it must increase when the reaction rate decreases'. It should be possible to take published data for a metabolic pathway, apply the Krebs-Kornberg principle and so determine which reaction is the pacemaker. It turns out that in some cases the Krebs-Kornberg principle cannot be applied. For example in some tissues when the rate of glycolysis is increased not a single compound shows the expected reduction in concentration.
12.1.2—
Occam's Razor
William of Occam, the mediaeval controversialist, formulated a procedural rule, that when a number of possible solutions can be proposed for a problem,
one should accept the simplest solution until it is shown to be untenable. The Krebs-Kornberg principle is a simple solution to a complex problem and in certain cases it yields a valid solution. However, when it is not applicable we must examine more complex solutions.
12.1.3—
Systems Properties
The rate equation for a metabolic pathway includes parameters for all the enzymes and variables for all the metabolites (Waley, 1964). The flux through the pathway is a systemic property in which all the parameters interact as a system. If a single parameter is altered, say the activity of a single enzyme, then the whole system responds and adjusts to that change. The extent to which an enzyme can be considered a control step is the extent to which a fractional change in its activity produces a fractional change in the flux through the system. The ratio
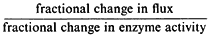
has been termed the sensitivity coefficient for that step (Kacser & Burns, 1973). If a 1 % change in activity of an enzyme produces a 1% change in flux through the system, then the sensitivity coefficient is 1 and the enzyme must be fully controlling the flux and is a pacemaker. However, the sum of all the sensitivity coefficients for a metabolic sequence is equal to unity and only in special circumstances can a single pacemaker be identified. That the sensitivity co-efficient of a particular enzyme is a system property, only in part determined by its own parameters, can be intuitively understood by an example. In a given situation an enzyme may have a very small sensitivity coefficient and contribute little to the overall control of the system. If however the activity of this enzyme is drastically reduced its sensitivity coefficient may be increased and it may contribute significantly to the overall flux. Since the sum of sensitivity coefficients must always equal unit the sensitivity coefficients of all the other enzymes must have changed despite the fact that their parameters are unchanged.
Much work on metabolic control is concerned with the identification of control points. Since each reaction contributes to the overall control a sense of judgement is necessary to identify major control points and it must be remembered that control may pass from one reaction to another as the flux through the system changes.
12.2—
The Identification of Control Points
12.2.1—
Equilibrium Considerations
Bücher and Russmann (1964) have proposed a model for metabolic control based on a controlled waterway (Fig. 12.1).
Figure 12.1
Schematic representation of a regulated waterway. The solid line represents the
situation when the flood gates are raised and there is little flow through the system.
The dashed line represents the increased flow when the flood gates are lowered.
The flow of water through the system is controlled by the flood gates. With a flood gate up, there is a negligible gradient across the reservoir and a slight flow of water over the dam. In metabolic terms the reaction
When the flood gate is lowered a slight gradient is produced across the reservoir and there is a rapid flow of water over the dam, which could be utilized to perform work. In metabolic terms, as the flux through the system increases, the reaction
12.2.2—
The Crossover Theorem
The crossover theorem states that: the variations of the concentrations of the metabolites before and after an enzyme which is a control point have different signs. It will be noted that this is only a formal statement of the situation at the waterfall in the Bücher and Russmann analogy, i.e. when the flux increases the concentration of the substrate falls and the concentration of the product
increases. However, the theorem is widely used, possibly because it presents data in a graphical form. Metabolic intermediates are plotted on the x coordinate in the sequence in which they are formed in the metabolic pathway and the concentration of each substrate, expressed as a percentage of the initial value is plotted on the y coordinate. An example is shown in Fig. 12.2 for the situation usually referred to as the 'Pasteur Effect'. Pasteur observed that 'oxygen inhibits fermentation' so that when carrot discs are transferred from air to nitrogen glycolysis increases with associated changes in the levels of intermediates. These changes are plotted for a period some 6 minutes after transfer from air to nitrogen (Fig. 12.2).
Figure 12.2
Crossover plot of glycolytic intermediates following transfer of
aged carrot discs from air to nitrogen (Data of Faiz-ur-Rahman,
Trewavas & Davies, 1974). Intermediates were measured before
transfer and 6 minutes after transfer from air to nitrogen.
The positive crossover observed between fructose-6-phosphate and fructose diphosphate suggests that phosphofructokinase is a control point. There is some confusion concerning the interpretation of negative crossovers. Some authors argue that a negative crossover does not indicate a control point. However, it must be remembered that when the flux through a system is changed, control can pass from one point to another. Thus in the case illustrated in Fig. 12.2 the increased flux through the glycolytic system involves an increase in activity of phosphofructokinase but with the increased rate of glycolysis, phosphoglycerate is removed more rapidly than it is formed, suggesting that a bottleneck exists between dihydroxyacetone phosphate and phosphoglycerate when phosphofructokinase is highly active.
The crossover theorem should be used with caution; the absence of a crossover should not be taken to mean the absence of a control point and there are many conditions which can produce crossovers indicating spurious control points. Before applying the crossover theorem, the reader is advised to consult the critique offered by Heinrich and Rapoport (1974) and to remember that
control is a property of the system; the identification of a control point is to focus attention on one component knowing that to varying degrees all other enzymes in the sequence contribute to the overall control.
12.3—
Control Mechanisms
Having identified possible control points it becomes necessary to determine the mechanism of control. Over a relatively long time scale control may be achieved by changing the amount of an enzyme but over a short time scale control is likely to be achieved by changing the activity of the enzyme.
12.3.1—
Control by Product Inhibition
Every enzyme has a certain affinity for its products and if the enzyme has a high affinity for its product then product inhibition will be pronounced. It appears likely that this form of control is restricted to some synthetic routes and to minor metabolic sequences.
12.3.2—
Control by Negative Feedback
The discovery of negative feedback in metabolic systems appears to have been made by Dische in 1940 who noted that 3-phosphoglycerate inhibits the phosphorylation of glucose in red cells and he proposed that this inhibition might play a regulatory function in glucose metabolism. His paper did not receive the attention it deserved and the idea of feedback had to be rediscovered.
In 1955 Roberts and his coworkers published the result of their extensive studies of the metabolism of E. coli grown on a medium containing 14 C-glucose. They found that the addition of one of a number of amino acids (known to be products of specific synthetic pathways) to the growth medium resulted in the virtually complete inhibition of incorporation of 14 C into that amino acid.
In 1956 Umbarger reported that isoleucine inhibited threonine deaminase—an enzyme which initiates the set of reactions leading to isoleucine biosynthesis. In the same year Yates and Pardee observed that cytidine monophosphate inhibits aspartate transcarbamylase—the first reaction of the pathway leading to the biosynthesis of the pyrimidines. These results offered a biochemical explanation for the end-product inhibition observed by Roberts with intact bacteria.
In 1959 Sir Hans Krebs introduced the first symposium to be held on metabolic control. Ideas on the regulation of carbohydrate and amino acid metabolism were discussed and the general ideas of feedback formulated. Control by negative feedback is an engineering concept and familiar examples include governers of steam engines, thermostats and automatic volume control in amplifiers. The limitations of the analogy between electrical feedback amplifiers and metabolic reactions have been stressed by Britton-Chance. However,
as with other terminology developed for use with metabolic control systems (see discussion of allosteric enzymes) the tendency has been to dispense with rigorous definitions and the general definition of negative feedback is 'increased output decreases the input'.
12.3.3—
Patterns of Control
The most frequently observed pattern of control is when the end. product of a metabolic sequence inhibits the first reaction belonging to that sequence. For example serine can be formed by the reaction sequence shown in Fig. 12.3.
Figure 12.3
Reactions involved in the biosynthesis of serine.
Serine inhibits 3-phosphoglycerate dehydrogenase and so controls its own biosynthesis. This simple mechanism is effective, presumably because a relatively small amount of the flux through the glycolytic pathway is directed towards serine and thus variations in the rate of serine production will not seriously perturb the glycolytic flux.
12.3.4—
Control of Branched Pathways
Inhibition of the first step in a metabolic pathway by an end-product leads to special problems in the case of branched biosynthetic pathways. Consider the sequence
It the first common step (A® B) is inhibited by either or both end-products, then an excess of one could inhibit the step A® B and lead to a deficiency of the
other end-product. Nature has evolved several mechanisms which avoid, to varying degrees, these difficulties. Stadtman (1970) has classified these mechanisms; here we discuss a few examples which have been studied in plants.
12.3.4.1—
The Aspartate Family
The formation of some amino acids from aspartate is outlined in Fig. 12.4 together with feedback controls which have been demonstrated.
Figure 12.4
Reactions involved in the biosynthesis of the aspartate family of amino acids.
® Single step reaction; ---® multi-step reaction; --® feed back loop.
Aspartokinase which catalyses the first reaction of the pathway is inhibited by lysine and threonine in a concerted or synergistic manner. The term concerted feedback inhibition has been used by Stadtman to describe cases of inhibition by two end products when the single end products produce no inhibition. The term synergistic inhibition is used when the total inhibition is much greater than the sum of their independent effects. The aspartokinase from plants appears to be inhibited by low concentrations of lysine but not by low concentrations of threonine—when both are present a much greater inhibition is observed. Although this effect would seem best to fit Stadtman's definition of synergistic inhibition it has been called concerted inhibition. Whatever the terminology, this mechanism does not involve methionine and is thus only a partial solution to the control of the branched pathway. The non-involvement of methionine means that in conditions where threonine and lysine
accumulate they will tend to inhibit the synthesis of methionine—even in situations where methionine may be in short supply. This imperfection is compounded by the feedback inhibition of homoserine dehydrogenase by threonine which further reduces the flow of intermediates available for the biosynthesis of methionine.
These imperfections in the control mechanism can be invoked to explain the effect of amino acids on the growth of Lemna minor (Table 12.1).
| ||||||||||||||||||||||||||||
The relatively small inhibition observed with lysine can be interpreted in terms of inhibition of aspartokinase (but see section on metabolic interlock ). The inhibition produced by threonine is consistent with the inhibition of homoserine dehydrogenase which would prevent the flow of carbon to methionine. The inhibition produced by homoserine is difficult to explain since as far as known it does not inhibit any of the enzymes involved. Furthermore in some plants e.g. peas, homoserine is produced in large quantities and transported in the phoem. The inhibition produced by isoleucine is consistent with a control pattern known as sequential inhibition. Isoleucine has been shown to inhibit threonine deaminase which would be expected to lead to an accumulation of threonine which in turn inhibits homoserine dehydrogenase and so reduces the flow of carbon to methionine. The inhibition produced by methionine is difficult to explain in terms of the reactions shown in Fig. 12.4. The pronounced inhibition observed with lysine and threonine is consistent with their synergistic effects on aspartokinase producing a reduction in the flow of carbon to methionine, which is reinforced by the inhibition of homoserine dehydrogenase
by threonine. Aspartic acid and isoleucine are unable to reduce the inhibition but methionine and homoserine are able to do so, presumably by supplying methionine directly or indirectly.
12.3.4.2—
Aromatic Biosynthesis
The control of aromatic amino acid biosynthesis has been extensively studied in fungi and bacteria and it is clear that nature has evolved many solutions to the problem of control in branched pathways. Some of the solutions which have been demonstrated in higher plants are shown in Fig. 12.5.
12.3.4.3—
Enzyme Multiplicity
In this control mechanism the first common step is catalysed by two or more enzymes which are under feedback control by compounds formed after the branching point. Four examples are given in Fig. 12.5, the control of chorismate mutase being particularly complicated. Three isozymes have been demonstrated in plants (Woodin & Nishioka, 1973); CM1 and CM3 are inhibited by phenylalanine and activated by tryptophane. CM1 and CM2 are inhibited by caffeic acid and chlorogenic acid whilst CM3 is inhibited by ferulic acid.
12.3.4.4—
Enzyme Aggregation
A number of fungi possess an aggregate of the five enzymes necessary for the conversion of 3-deoxy-D -heptulosonate-7-phosphate to enolpyruvyl-shikimate-5-phosphate. In Neurospora the five enzymes form a complex (M.W. 230,000) coded by the arom gene cluster. This large complex has not been demonstrated in higher plants, but a bifunctional enzyme consisting of dehydroquinase and shikimate dehydrogenase has been isolated (Boudet, 1971). Neurospora contains two dehydroquinases, the one in the aggregate is involved in an anabolic sequence, the other maps outside the arom cluster and is thought to function as a component of a catabolic sequence. The physiological significance of the multienzyme cluster could be to separate the degradative and synthetic routes. This could be achieved if the dehydroquinic acid which is formed in the aggregate is not released as a free product. This idea has become known as metabolic channelling and in a number of cases it has been shown that metabolites involved in channelling do not leave the enzyme surface. For example fungi possess a bifunctional enzyme consisting of carbamyl phosphate synthetase and aspartate transcarbamylase.
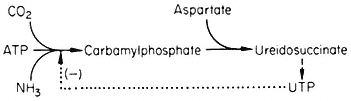
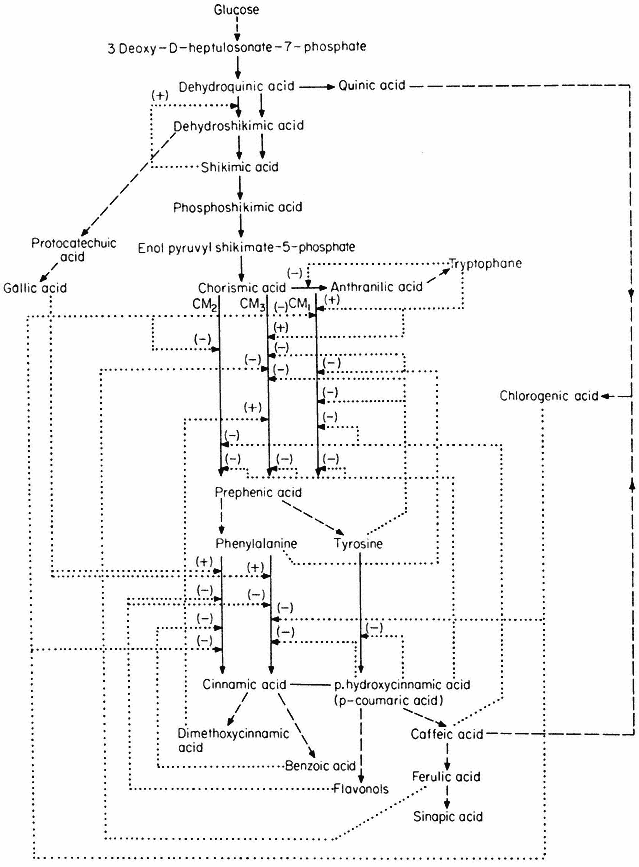
Figure 12.5
Reactions involved in the biosynthesis of aromatic compounds.
® Single step reactions; --® multi-step reactions; --® feed back loop.
When the bifunctional enzyme is synthesizing ureidosuccinate from 14 CO2 , the addition of carbamyl phosphate does not dilute the incorporation of 14 C into ureidosuccinate, showing that carbamyl phosphate formed by the double enzyme does not equilibrate with free carbamyl phosphate.
Higher plants contain two dehydroquinases one forming a bifunctional association with shikimate dehydrogenase, the other being activated by shikimic
acid. Such a system involving a specific regulation of isoenzymes and molecular compartmentalization may function to control the partitioning of dehydroquinic acid between the pathways to phenolcarboxylic acids and aromatic amino acids.
12.3.5—
Metabolic Interlock
The reactions and feedback loops shown in Fig. 12.5 give some idea of the complexity of control within a metabolic pathway. When the feedback loops of other metabolic pathways are taken into consideration it seems highly likely that the control must be interlocked. As an example, consider the reactions involved in the biosynthesis of methionine. In Fig. 12.4, this is shown as the addition of a C1 group to homocysteine. The synthesis of this C1 group is shown in Fig. 12.6.
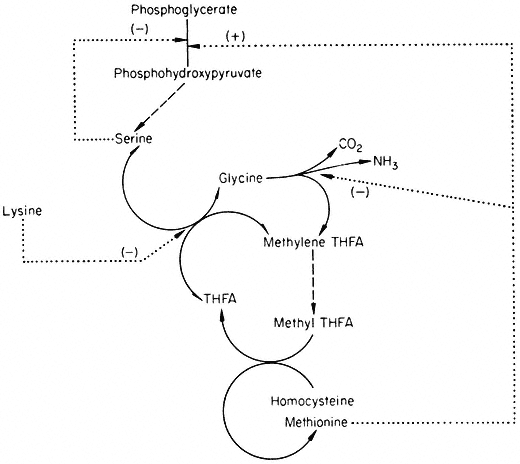
Figure 12.6
Reactions involved in the biosynthesis of methionine.
® Single step reaction; ---® multi-step reaction; . . .® feed back loop.
The active C1 group is generated in the serine hydroxymethyl transferase reaction as methylene tetrahydrofolate. After reduction to methyl THFA the methyl group is transferred to homocysteine to form methionine. Homocysteine is derived from the aspartate family (cf. Fig. 12.4) and so is lysine. The metabolism of the aspartate family and the serine glycine family are thus interlocked via lysine, which inhibits both aspartokinase and serine hydroxymethyl transferase.
12.3.6—
Enzymes as Control Elements
Until the discovery of feedback inhibition in 1956 enzymes were considered as biological catalysts and most enzymologists were concerned with the properties of the active site. When it became apparent that enzyme activity was under the control of specific metabolites, which were structurally unlike the active site, it became necessary to postulate a control site to which metabolites could bind and modulate the activity of the enzyme. Recognizing the lack of structural similarity between the substrate and the effector the term allosteric site was introduced (Monod, Changeux & Jacob, 1963). Subsequently Monod, Wyman and Changeux (1965) noted that many regulatory enzymes with specific allosteric sites exhibit cooperative-type kinetics and they developed a concerted transition hypothesis to explain the observed kinetics. Many workers assume that any enzyme showing sigmoidal kinetics is an allosteric enzyme and some workers imply that the mechanism of activation of all allosteric enzymes follows the mechanism postulated by Monod et al., (1965). The purist may object but the original precise definition has been replaced by a broader but less precise usage.
12.3.6.1—
Allosteric Enzymes
Before 1956, the kinetics of numerous enzymes were shown to follow the rate law of Michaelis and Menten giving a hyperbolic plot of v against S . An examination of enzymes which were known to be subject to feedback regulation showed that in some cases the relationship between v and S was sigmoid. In other cases the addition of a feedback inhibitor led to a change from a hyperbolic to a sigmoid relationship.
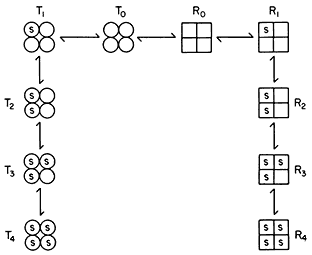
Figure 12.7
Illustration of the Monod model of an allosteric enzyme.
Monod et al., (1965) proposed a simple model to account for the observed kinetics in which they took account of the observation that allosteric enzymes are composed of subunits. They assumed that the protein subunits were arranged in such a way that they occupy equivalent positions so that the enzyme must have at least one axis of symmetry. They further assumed two interconvertible states—R and T—each maintaining the symmetry principle. A simple illustration of the model is given in Fig. 12.7 based on the symbols used by Koshland (1970). The reader is referred to the original article by Monod et al., (1965) for the derivation of the rate law. However a consideration of the illustration leads to a basic understanding of the underlying causes of sigmoid kinetics. Two types of allosteric behaviour may be distinguished—one the variable K system in which the two states of the enzyme have different affinities for the substrate, the other, the variable V system in which the R state is catalytically more active (Fig. 12.8).
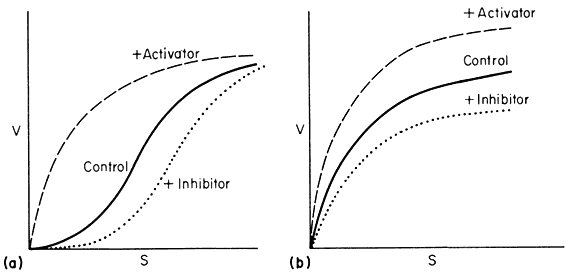
Figure 12.8
Relationship between v and s.
(A) a variable K system; (B) a variable V system.
Variable K Systems
In the variable K system, the R form is assumed to have a higher affinity for S than the T form of the enzyme. If in the absence of ligands, the equilibrium favours the T form then increasing concentrations of S will shift the equilibrium towards the R state giving sigmoid v against S plots. Another way of looking at this situation is to imagine that the interconversion of the R and T forms can be blocked and the enzyme isolated as the R or T form. The T form would give a normal Michaelis-Menten curve with a high Km(T ) and the R form would also give a Michaelis-Menten curve but with a low Km(R ) . Allowing R and T to be interconvertible allows Km to vary.
If an activator binds preferentially to the R state it will tend to put the enzyme entirely in the R state and so remove the sigmoidicity of the v against S plot. Similarly if an inhibitor binds preferentially to the T form it will increase the sigmoidicity.
Variable V Systems
In the variable V system the R form is assumed to be catalytically more active than the T form though both forms have the same affinity for the substrate so that the v/S plot is hyperbolic in the presence or absence of activators and inhibitors. An activator is assumed to have a greater affinity for the R form than for the T form whilst the reverse holds for an inhibitor.
12.3.6.2—
Alternative Models for Allosteric Enzymes
This superficial consideration of the Monod model concentrates on only one aspect of the model–the two configurational states. This limited aspect gives a qualitative explanation of' sigmoid kinetics described by the equation:
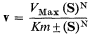
where N = 2.
To explain higher order reactions the Monod model includes some highly restrictive assumptions which produce mathematical simplicity but are nevertheless based on the fundamental properties of protein structure.
An alternative model proposed by Koshland (1970) assumes an induced fit so that the protein subunit undergoes a conformational change when it binds a ligand. A simple sequential model may be illustrated:
This simple model is not a necessary part of the induced fit model (which in fact suggests a diversity of forms of association between enzyme and ligand—25 for a tetrameric protein). Nevertheless it simplifies the mathematical treatment and in many cases provides a close fit to experimental values.
12.3.6.3—
Kinetic Constants for Allosteric Enzymes
When an electronic engineer constructs a control circuit he needs to know the characteristics of the components—the amplification factor, grid bias, etc. Similarly if we wish to understand the control of a system we need to know the characteristics of its components, that is, the kinetic constants of the enzymes. Clearly a variable K system cannot be defined by a Km value nevertheless the same operation value can be used, that is the concentration of substrate producing half maximum velocity which is in this case designated S0.5 . More generally the term (X0.5 ) is used to designate the ligand concentration at half saturation.
The constant N is operationally the order of the reaction, its physical meaning may relate to the number of substrate binding sites on the enzyme or to the degree of cooperativity of binding. Whatever the meaning of N it can be estimated by means of a Hill plot (Fig. 12.9). Another kinetic term, Rs is the ratio between the 90% and 10% saturation values. Rs will be 81 for all curves following Michaelis-Menten kinetics and less than 81 for cases of positive cooperativity–a few cases of negative cooperativity are known and these give Rs values greater than 81.
Figure 12.9
Kinetic data for an enzyme with variable 'K' properties.
The curves are drawn assuming
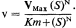
Valve characteristics define the properties of the valve but do not determine the function of the valve–the function of the valve is determined by the circuit in which it operates, in one circuit it may function as a cathode follower, in another as an amplifier. Similarly, the kinetic constants of a control enzyme do not determine its role in metabolic organization. The properties of an enzyme in a metabolic pathway are modified by its association with other enzymes. The study of control in metabolic pathways relates enzymology to physiology and illustrates the somewhat unfashionable view that the whole is more than the sum of the parts.
12.4—
Control Mechanisms Involving Changes in the Amount of Enzyme
The brilliant work of Jacob and Monod (1961) on the regulatory mechanisms governing b -galactosidase activity in E. coli led to many attempts to demonstrate similar control systems in plants. A long series of negative experiments has made it unlikely that operons exist in higher plants. There is, however, evidence for the interdependent regulation of some groups of enzymes. For example in parsley and soybean cell cultures, the three enzymes catalysing the synthesis of p -coumaroyl CoA from phenylalanine show strictly concomitant changes in activity. Large changes in activity are initiated by light but there is no satisfactory explanation for the strict coordination in activity. Mohr (1972) has suggested that genes can be divided into four classes—active genes which function in the same way in light and dark, inactive genes which function only at special times in the life history of the plant (e.g. flowering genes), potentially active genes whose activation is under the control of the phytochrome system and repressible genes which can be repressed by the phytochrome system. Positive photoresponses are identified with gene activation and negative photoresponses with gene repression. This model is so basic that it must contain some truth, but it offers no explanation of the mechanism of regulation other than to identify regulation as occurring at the gene level. Much of the evidence for Mohr's model is concerned with the increase in phenylalanine ammonia lyase which occurs when mustard cotyledons are exposed to far-red light. Recently however, it has been suggested (Attridge et al., 1974) that this increase in activity is due to an activation of pre-existing enzyme rather than synthesis.
Following the success in understanding the regulatory system in prokaryotes it was clearly sensible to see if the system was applicable to plants. It now appears that the regulatory systems are very different, but the technical vocabulary used in the study of prokaryotes is still used in studies with plants. It should be noted that terms such as repression and derepression have precise meanings in relation to molecular models of regulation in prokaryotes, but when applied to plants, these terms should not be taken to imply a mechanism similar to that found in bacteria.
Many cases are known where a metabolite reduces the level of an enzyme involved in its metabolism (Table 12.2). Most of these cases are reported as
| |||||||||||||||||||||
examples of repression but the mechanism involved varies from case to case—for example arginine probably exerts its effect by stimulating proteolysis, the effect of ammonia may be due to proteolysis, though the formation of a specific protein inactivator has been proposed, and the effect of glucose on invertase has been reported as due to the destruction of m-RNA required for invertase synthesis.
The regulation of enzyme levels may involve one or more of the following:
(a) Degradation
(b) Inactivation
(c) Activation
(d) Synthesis
When examining a particular system it is necessary to establish which control system is involved and to measure its rate—this is frequently surprisingly difficult.
12.4.1—
Protein Turnover and Degradation
The view that plant proteins may undergo a cycle of synthesis and degradation was first proposed by Gregory and Sen in 1937. The terminology is somewhat confused; protein turnover implies synthesis and degradation but usually only degradation is measured.
Three methods are available for determining the overall rate of protein degradation.
(a) Incorporation of 14 C-amino acids.
The tissue is pulse-labelled with a 14 C-amino acid and the incorporation into protein measured. During the chase period the rate of protein degradation is estimated from the fall of radioactivity in the protein. The method has two serious disadvantages: (1) The recycling of protein amino acids—that is the reincorporation of a 14 C-amino acid released by degradation tends to underestimate the rate of degradation. This disadvantage can be minimized by using a 14 C-amino acid which is rapidly metabolized; (2) The existence of pools of amino acids means that the specific activity of the amino acid at the site of protein synthesis is unknown. Furthermore the incorporation of a labelled amino acid into a metabolic pool during the pulse period may be followed by a slow release of 14 C-amino acid during the chase. This disadvantage can be minimized by using an amino acid whose pool size is small, e.g. leucine.
(b) Measurement of the specific activity of aminoacyl t-RNA.
Trewavas (1969) has developed a method in which the specific activity of the amino acid entering protein during a labelling experiment is determined by isolating the appropriate aminoacyl t-RNA.
(c) Measurement of the incorporation of 3 H.
Humphrey and Davies (1975) established that when Lemna fronds are exposed to 3 H2 O, the tritium rapidly enters and equilibrates with H on the a -carbon atoms of amino acids. Protein synthesized during the exposure to 3 H2 O is thus
labelled in a stable position. When the plants are transferred to H2 O the amino acids rapidly lose their 3 H, the 3 H in protein is stable, but as soon as the protein is degraded to amino acids the 3 H exchanges with H2 O. Thus the rate of protein degradation can be determined simply by measuring the loss of 3 H from protein. The method depends upon the speed with which transaminases catalyse the exchange reaction between water and the hydrogen on the a -carbon atom of amino acids. If it can be established for a particular tissue that this reaction is fast relative to the rate of protein synthesis the method overcomes problems of pools and recycling. The method can in principle be applied to a specific enzyme, but this requires the isolation of the pure enzyme.
12.4.1.1—
Degradation of Specific Enzymes
If the degradation of proteins were a random process then a low rate of protein hydrolysis would almost completely destroy overall enzyme activity. This is because a 5% per day rate of degradation means that each enzyme molecule would be subject to an average degradation of 5% and for many enzymes this would mean a complete loss of catalytic activity. Consequently we must look for specificity in degradation.
A NADH-nitrate reductase-inactivating enzyme has been isolated from maize roots which appears to have a high degree of specificity (Wallace, 1974). The enzyme has been purified 460-fold, the purified preparation has proteolytic activity but no peptidase activity and the capacity to inactivate nitrate reductase is blocked by phenylmethylsulphonyl fluoride suggesting the involvement of a serine residue at the active site.
12.4.1.2—
The Measurement of Enzyme Half-Lives
If protein synthesis is blocked by an appropriate inhibitor and at various time intervals the level of enzyme is measured, we can determine the fall in activity of the enzyme. If we assume that the inhibitor completely blocks protein synthesis without interfering with the reactions involved in inactivating or degrading the enzyme we can estimate its half-life (Table 12.3).
| ||||||||||||||||||||||||||||||
This method is open to criticism since in a number of cases inhibitors of protein synthesis have been shown to interfere with enzyme degradation. Thus cycloheximide stops the decline in phenylalanine ammonia lyase which follows the light induced elevation of the enzyme in potato discs and also the decline in phosphoglucomutase which occurs on slicing potato tubers. In these cases, cycloheximide is assumed to block the synthesis of the specific proteolytic enzymes necessary for the removal of the lyase and the mutase.
12.4.2—
Enzyme Inactivation
Experimentally it is difficult to distinguish between enzyme degradation and inactivation. This is reflected in the terminology, thus the enzyme described in the previous section which is thought to degrade nitrate reductase is referred to as a nitrate reductase inactivating enzyme by Wallace. A simple kinetic test enables an investigator to determine if enzyme activity is changing during the period of an assay. The rate equation for an enzyme catalysed reaction can be written:
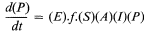
Provided that (S ) substrate, (A ) activator and (I ) inhibitor are constant in a series of assays, the integral form of the equation is
Thus for a series of assays when the amount of product formed is plotted not against time, but against time multiplied by enzyme concentration, all the values should fall on a single curve. If the points do not fall on a single curve then it is likely that the enzyme is being degraded or inactivated in some way (Selwyn, 1964).
12.4.2.1—
Inactivation by Protein-Protein Interaction
The modulation of enzyme activity by the interaction of protein subunits is widely recognized. A number of reports suggest that interaction between different proteins may result in enzyme inactivation. For example the naturally occurring inhibitor of potato tuber invertase appears to be a protein which forms an essentially undissociable complex with invertase. The inactivation of Lemna nitrate reductase has been attributed to protein-protein interaction and ammonium adapted plants are said to contain a protein which inhibits nitrate reductase.
However, the most clearly established case is the interaction between ornithine transcarbamylase and arginase in Saccharomyces (Wiame et al., 1973). The activity of ornithine transcarbamylase was shown to decline following the addition of arginase. It was established that arginase binds to the ornithine
transcarbamylase and inhibits its activity. The binding is on a one to one basis and does not reduce the arginase activity. This interaction has not been observed in other organisms.
12.4.2.2—
Chemical Modification
A number of enzymes are inactivated by low concentrations of pyridoxal phosphate at physiological pH. The mechanism of inactivation is the formation of a Schiff base between pyridoxal phosphate and an amino group of the protein, usually the e -amino group of lysine.
Glutamate dehydrogenase and aldolase have been shown to be inactivated by pyridoxal phosphate and to be protected by their substrates, suggesting that lysine residues are involved in the active centre of these enzymes. It is difficult to assess the physiological significance of these reactions; the concentration of free pyridoxal phosphate in plants is low but the ratio pyridoxal phosphate: pyridoxamine phosphate changes with the nitrogen status of the plant and the possibility of this ratio monitoring the nitrogen status of the plant and controlling the activities of key enzyme has been discussed.
Carbamyl phosphate also reacts with e -amino groups of lysine, the rate of carbamylation varying from protein to protein. Glutamate dehydrogenase is readily inactivated by carbamyl phosphate but it is doubtful that carbamylation of enzymes has physiological significance in plants.
Enzyme catalysed chemical modification is well documented in animals and bacteria and among the important examples we note glycogen phosphorylase and glycogen synthetase of E. coli which is adenylated by ATP. Adenylation has not been observed in plants but phosphorylation, methylation and acetylation of proteins has been demonstrated. The extent to which these modifications are important regulatory mechanisms remains to be determined.
The classification of chemically modified enzymes in terms of activation or inactivation is somewhat artificial. For example phosphorylation leads to an activation of glycogen phosphorylase but an inactivation of glycogen synthetase. These chemical modifications could equally well be considered as dephosphorylation in which case glycogen phosphorylase is inactivated and glycogen synthetase activated. However, from an operational point of view, whether an enzyme is activated or inactivated is of paramount importance.
12.4.3—
Enzyme Activation
Four enzymes of the Calvin cycle and two enzymes of the Hatch-Slack pathway are light activated whilst glucose-6-phosphate dehydrogenase, an enzyme of the pentose phosphate pathway is light inactivated. It is highly probable that a number of these responses reflect a reductive process, since dithiothreitol treatment of crude extracts gives results similar to light treatment of intact seedlings. However, in some cases the reaction is certainly more complicated
than a direct response to a change in the SH/SS redox potential. A scheme for the activation/inactivation of pyruvate phosphate dikinase is shown in Fig. 12.10.
Figure 12.10
Schematic representation of the processes of activation and inactivation of
pyruvate, P: dikinase. The process leading to the irreversible inactivation
of both active and inactive enzyme in vitro apparently include an O2
-dependent oxidative reaction involving thiol groups.
(After Hatch & Slack, 1969.)
The mechanism of activation may vary from species to species. Thus the light-activation of pea NADP glyceraldehyde phosphate dehydrogenase appears to involve a conformational change produced by reduction and leads to an increase in Vmax . The spinach enzyme however, appears to need NADPH or NADP or ATP in addition to a reducing agent for activation. The activation involves dissociation of a tetramer leading to an increased affinity for NADP.
An interesting suggestion for the blue light activation of phenylalanine ammonia lyase has been proposed by Engelsma (1974). He suggests that the enzyme is present in gherkin hypocotyls in an inactive form due to inhibition by trans-hydroxycinnamic acid. Blue light (in the presence of a photoreceptor such as riboflavin) converts the trans- to the cis-hydroxycinnamic acid which is much less inhibitory. A phytochrome-mediated activation of phenylalanine ammonia lyase has also been reported in mustard cotyledons by Attridge et al., (1974). It is not clear if the cis-trans isomerization is applicable to the phytochrome-mediated activation which is inhibited by cycloheximide.
12.4.4—
Enzyme Synthesis
In the absence of activation or inactivation, the net synthesis of an enzyme is given by gross synthesis minus degradation. Net synthesis is measured as the change of activity so that gross synthesis can be measured if the rate of degradation is known. Because of the technical difficulties involved in obtaining quantitative data most workers have been concerned to develop methods which establish that the increase in activity of an enzyme is due to synthesis rather than degradation.
12.4.4.1—
Density Labelling
Gibberellic acid stimulates the synthesis of a number of enzymes in the aleurone layer of barley seeds. Filner and Varner (1967) introduced a method of demonstrating that the bulk of a -amylase which appears after treatment with gibberellic acid is due to protein synthesis. They incubated aleurone cells with H218 O so that when storage proteins were hydrolysed 18 O-labelled amino acids were formed.
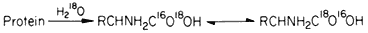
When new protein is synthesized from the 18 O-labelled amino acids it will have a greater density due to the incorporation of 18 O into the peptide links. The density difference between 18 O and 16 O containing proteins allows separation by equilibrium density gradient centrifugation. The observed increase in density was consistent with the bulk of the new a -amylase being synthesized from amino acids. A major limitation is that proteolysis is required to introduce 18 O into amino acids and this restricts the method to studies of seed germination.
An alternative method is to introduce 2 H into the protein. This can be achieved by incubating a tissue in 2 H2 O so that 2 H labels the amino acids by entering at the a -carbon atom in a transaminase catlysed exchange reaction. With prolonged incubation in 2 H2 O, deuterium enters the amino acids at other points producing large changes in the density of newly formed protein.
The major advantage of the density labelling methods is that they enable the synthesis of a particular enzyme to be demonstrated without purifying the enzyme. However, the method requires expensive equipment and is time consuming. Consequently many investigators rely on data obtained by the use of inhibitors of protein synthesis. Such data is rapidly obtained and at low cost but its interpretation must always be equivocal.
12.4.4.2—
Enzyme Induction
Filner et al., (1969) have listed the numerous examples of increases in enzyme activity in response to age, hormonal or environmental changes. In many cases these increases may be due to enzyme synthesis, though the evidence is often little more than that gained by the use of inhibitors of protein synthesis. The evidence for substrate-induced enzyme synthesis is extensive in the case of nitrate reductase but in other cases, for example the induction of a specific isoenzyme of glutamate dehydrogenase, the case is not yet proven.
12.5—
Final Comments
The basic similarity of living organisms is reflected in their biochemistry, the major metabolic pathways being common to all living organisms. The diversity of living organisms is reflected in their specialized control systems. The control
mechanisms of one species may be very different from those of a closely related species. The investigator has a daunting task when he considers the amount of work necessary to establish control mechanisms over a wide range of species.
The difficulties of comprehending control mechanisms even in a single organism are no less daunting. The difficulties stem from the number of assays necessary to establish the complete rate law for a regulatory enzyme. For example the enzyme glutamine synthetase from E. coli is known to be affected by eight reactants and modifiers. Hence if 6 points per curve are necessary for a kinetic analysis, then to establish the rate law for glutamine synthetase will require in excess of 1.6 × 106 assays. The task of analysing the many enzymes involved in metabolism is frightening and when we recognize that all the enzymes interact and affect one another the system defies analysis even with modern computers.
The way ahead seems to require inspired guesses at the nature of the control mechanisms involved, followed by model building and a systems analysis—probably making linear approximations—to see if the behaviour of the biological system can be predicted.
The difficulty in analysing control systems may be compared with the difficulties in understanding economics. The main control step of glycolysis appears to be phosphofructokinase, a reaction which involves a loss of free energy of 4 Kcals per mole. This loss may be considered the cost of controlling glycolysis. Protein degradation involves a large loss of free energy and this may be the price the cell has to pay for its complicated but effective control mechanisms. The situation can be likened to the economy of surplus production in society—in the Brave New World of Aldous Huxley products were destroyed to maintain the economy—perhaps nature is a capitalist!
Further Reading
Bayer P.D. (1970) (Ed.) The Enzymes. See articles by Koshland D.E., Stadtman E.R., & Atkinson D.E. Academic Press.
Cohen G.N. (1968) The Regulation of Cell Metabolism. Holt, Rhinehart & Winston.
Davies D.D. (1973) (Ed.) Rate control of biological processes. Symposia of the Society for Experimental Biology. Vol 27.
Kun E. & Grisolia S. (Eds.) (1972) Biochemical Regulatory Mechanisms in Eukaryotic Cells. Wiley & Sons.
Milborrow B.V. (Ed.) (1973) Biosynthesis and its Control in Plants. Phytochem. Soc. Sym. Series No. 9.
Newsholme E.A. & Start C. (1973) Regulation in Metabolism. Wiley & Sons.
Chapter 13—
Hormone Action
13.1—
Introduction
The genesis of hormone research in plants occurred with the discoveries of Charles Darwin and his son Francis, who published, almost a century ago, their book entitled The Power of Movement in Plants'. Their experiments dealing with phototropism revealed that an 'influence' produced in the tip of a canary grass coleoptile was translocated to the basal portion where it caused the organ to grow towards the light. Almost fifty years later Went isolated a substance with just these properties which was produced in the tip of coleoptiles and transported to the basal portion of the organ. This substance was named auxin. Ten years after Went's (1934) discovery, the structure of this material was identified as indoleacetic acid (IAA).
The nature of auxin effects on plant growth and development was studied in many laboratories in the late 1930's and 1940's. However, it was not until World War II that the selective action of indoleacetic acid served as a basis for developing new chemicals for the control of growth in specific plants. At that time phenoxyacetic acids were discovered and their derivatives were carefully studied under the cover of military secrecy, both in the United States and England. The results of those studies were first published in scientific journals in 1945 and 1947. Today the most widespread uses of growth regulators are as herbicides.
Plant hormones may be divided into five general groups. Auxins include the native indoleacetic acid, as well as the synthetic phenoxyacetic acids, notably 2,4-D (2,4-dichlorophenoxyacetic acid). The second group is the gibberellins which are steroid-like compounds comprising over forty different structures. The third group, the cytokinins, are all N 6 substituted adenosine compounds. The fourth class is represented by a single substance, abscisic acid, which is also derived from isoprenoid units as are the gibberellins. Ethylene, the simplest of all the plant hormones, is a gas and therefore is easily spread from organs of production to organs of sensitivity.
It is interesting that of these five classes of hormones, two are generated from mevalonic acid. Ethylene is produced by the metabolism of methionine and indoleacetic acid is produced by the removal of carbon and nitrogen from tryptophane. Cytokinins, in terms of synthesis, are probably the most complex hormones. They are possibly produced from the breakdown of transfer RNA (tRNA). In the intact tRNA, the adenosine moiety of the RNA is modified by the addition of an isoprenoid group; subsequently, the tRNA is probably degraded, yielding isopentenyl adenosine, (IPA) the endogenous cytokinin.
One might reasonably imagine that an understanding of the physiological role and biochemical mechanisms of action of the plant hormones would have been elucidated quite some time ago. This of course, is in relation to what is known about the many and complex hormones in animals. Unfortunately, however, the plant hormones are not as well understood. Probably the main obstacle to the elucidation of the action of plant hormones has been the fact that they have overlapping and complementary effects on the actions of each other.
During the years between 1960 and 1970 substantial progress appeared to be made on the biochemical mechanisms of action of plant hormones. Much of this research dealt with the effect of the hormones on nucleic acid metabolism, protein synthesis and the synthesis of specific enzymes, since it was felt that the mechanism of hormone action would involve the control of the production of messenger-RNA (mRNA) and the de novo synthesis of enzymes. In the late 1960's the general nature of the research was radically changed because many investigators had found that the plant hormones mediated an effect on cell growth within a few minutes. This information implied a very rapid primary response mechanism of hormone action; consequently, these effects could not be mediated by changing the rate or type of synthesis of nucleic acids or of specific proteins. Many years of research have since been devoted to studies dealing with short-time growth responses to plant hormones. It now seems most likely that plant hormones regulate plant growth through both short-term and long-term growth controls.
It is fairly well understood that animal hormones first interact with the animal cell by binding to, or reacting with, a receptor site of some type within the target cell. Plant physiologists and biochemists are now beginning to consider that the action of many of the plant hormones similarly involves their binding to receptor agents which in turn amplify the action of the hormone and thereby bring about specific changes in nucleic acid synthesis, protein synthesis, enzyme activity and possibly other physiological responses, such as changes in permeability of membranes.
This chapter will summarize the historical evidence which has led to the current level of understanding of the action of plant hormones. Emphasis is placed on the reactive sites within target cells and how the interaction of the hormones with their specific receptors may regulate the growth and development of plant cells.
When a hormone is applied to a responsive plant system it brings about a specific change which results eventually in a measurable biochemical or physiological effect. Two distinct aspects of the measured effect are involved: the specific change in metabolism and the series of steps which lead to the physiological effect. Usually, the molecular interaction of the hormone at its site of action is referred to as the mechanism of action. The subsequent sequence of reactions leading to the physiological effect is referred to as the mode of action. Therefore, by definition each hormone has its own distinctive mechanism of
action even though the manifestation of the hormone mechanism may depend upon prior action by other factors. Thus, it is possible for the hormonal mechanisms in one plant system to lead to a series of physiological responses which may be completely different from those in a second system. This difference in mode of action can be brought about because the second system has more or less of another hormone, has other biochemically rate-limiting components or possesses structural and cytological differences.
13.2—
Auxin Actions
13.2.1—
Introduction
Amongst plant growth regulators, the auxins have been studied the longest and have involved the greatest amount of research. This class of hormones is known to promote cell enlargement or cell elongation, a process which requires extension of the cell wall. Figure 13.1 shows the chemical structures of the major natural and synthetic auxins.
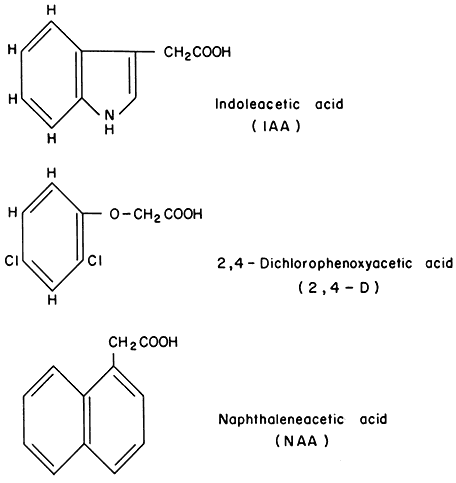
Figure 13.1
Structure of various auxins.
Commencing in 1940, the majority of studies involving auxins were begun in an attempt to determine the molecular interaction with the plant cell. Most of this research was directed toward the cell wall because it was believed that auxin
action required a change in the cell wall to allow for cellular expansion. Thus, the effects of the hormone were thought either to change the cell wall deposition or to hydrolyse certain cross-linkages in cell walls making them more elastic.
This type of research was carried out for several years without achieving a clear understanding of how auxin might control such processes within the cell wall. In the 1950's a new area of research was begun. Under the direction of F. K. Skoog in Madison, Wisconsin, a report (Silberger & Skoog, 1953) was published which showed that the auxin, indole-3-acetic acid, remarkedly affected the RNA and DNA contents of plants. Auxin increased the content of nucleic acids in tobacco tissue cultured on a sucrose-agar medium. This increase occurred prior to the auxin-induced growth of the tissue at concentrations of IAA which were optimal for cell enlargement. For many years following this discovery much research was devoted to the general mechanism by which auxin increased the synthesis of nucleic acids.
A great share of this research was begun in J. Hanson's laboratory in Urbana, Illinois by West and Key (West et al., 1960) who showed that the synthetic auxin, 2,4-D, (see (Fig. 13.1), produced a wide range of morphological and physiological changes in the hypocotyl of soybean and the mesocotyl of corn.
Within 15–24 hours after 2,4-D treatment, cellular enlargement was noted concomitant with an increase in the size of the nucleus. Accompanying these changes were dramatic increases in RNA content, most of which were due to increased ribosomal-RNA. Chrispeels and Hanson (1962) suggested that auxin acts on the nucleus causing it to revert to a meristematic type of metabolism. The role of the nucleus in such a sequence of events is of obvious importance since the nucleus has been shown to accumulate RNA in response to auxin. Beginning in the 1960's, Key began experiments on auxin regulation of nucleic acid synthesis. He and Ingle (1964) demonstrated that auxin controls the synthesis of nucleic acids other than that of ribosomal-RNA. Their data revealed that auxin causes production of nucleic acid which appears to be of the messenger-RNA type.
Subsequent experiments by O'Brien et al., (1968) showed that treatment of soybean with auxin caused a large increase in chromatin-directed RNA synthesis. It was of interest to note that auxin-induced RNA synthesis produced a type of RNA which was different from the control RNA as judged by molecular size and nearest neighbour analysis. Following this area of research other studies (Hardin & Cherry, 1972; Hardin et al., 1970; Hardin et al., 1972) showed that 2,4-D increased the activity of RNA polymerase. Current research suggests that particular cytoplasmic or membrane bound factors may enhance the activity of RNA polymerase. It is hypothesized (Hardin et al., 1972) that the mechanism of action of auxin is to bind to a receptor molecule located on the plasma membrane. The receptor is then released and moves into the nucleus where it modulates the activity of the RNA polymerase. The increase in RNA polymerase activity leads to increased synthesis of messenger-RNAs, which in turn regulate, or control, the synthesis of specific proteins within the target cells.
A primary challenge is to isolate the factor which binds to, or reacts with, auxin, and determine whether this is the primary action of the hormone. Secondarily, research is needed to determine what series of events this interaction puts into metabolic play. It is likely, once the auxin has bound to the receptor molecule within the target cells, that the changes in nucleic acid synthesis cause an increased activity of the various enzymes associated with the cell wall. Many other activities which have been measured over the many years are probably secondary effects resulting from the primary action of the auxin reacting with its receptor molecule.
13.2.2—
Regulation of Cell Wall Extension Ability
When plant cell walls elongate, an accompanying increase in weight and volume also takes place. This increase in size requires that the cell wall be increased in mass as well as area. Even though the dry weight of wall material greatly increases due to cellular enlargement, the thickness and density of the wall remains constant. Thus, wall synthesis appears to be a fundamental requirement during cell elongation. In addition, an increase in wall mass must come from a deposition of polysaccharides. It was believed for many years that auxin, in this particular case indoleacetic acid, increased cell elongation by promoting cell wall synthesis. Auxin, in order to allow for the increased wall extensibility, may also regulate the activity of enzymes involved in cell wall loosening which catalyse the breaking of various cross-linkages between the wall microfibrils.
Plant tissues which respond to auxin by an increased rate of cellular elongation also exhibit an increase in cell wall loosening. However, the kinetics of auxin-induced wall loosening vary considerably from tissue to tissue. For example, in dark grown maize coleoptiles wall loosening is induced by concentrations of auxin around 10–4M , and the increase in loosening proceeds very slowly with time. However, the same tissue responds to auxin very dramatically as maximal increases in total length occur between two and six hours after exposure to auxin. It is of interest, therefore, that the rate of cell wall elongation, as measured by increased section length, is much greater than the increase in cell wall extensibility.
Thus, considerable changes in cell wall extensibility in a given tissue as caused by auxin, may affect growth in a different manner. In certain tissues the rate of growth is constant whilst total wall extensibility changes. In other tissues wall extensibility remains constant during a period of changing growth rate. At the present time it seems that auxin causes a biochemical change in the wall probably by breaking or modifying the cross-links between the wall polysaccharide chains. These changes in the cell wall are then translated under cell turgor pressure into wall elongation. It is now relevant to discuss how auxin affects the cross-links in a cell wall and what the biochemical changes are which take place in a cell wall during cell elongation. In this regard, it is necessary to look at the effect of auxin on various enzymes associated with the cell wall in the context of how these enzymes affect cell wall loosening properties.
13.2.3—
Action on Cell Wall Associated Enzymes
The most dramatic auxin effect on enzyme formation is the induction of cellulase, polygalacturonase and other hydrolases in pea cotyledons treated with a high concentration of IAA (Datko & Maclachlan, 1968). Detection of in vitro formation of cellulase by a ribosomal preparation from peas has been claimed. This was reported to be enhanced using a ribosomal system prepared from IAA-treated peas (Davis & Maclachlan, 1969). The large IAA effect on cellulase formation in vivo is a slow response occurring over several days and cannot be involved in a rapid auxin action on elongation. However, it could be a cause of the lateral swelling response seen in pea cells following IAA treatment.
Auxin stimulates not only the synthesis of cellulase but also the level of glucan synthetase. Masuda (1968) and Masuda and Yamamoto (1970) found that the fungal b -1,3-glucanase, isolated from cultures of Sclerotinia libertans induced rapid elongation of excised oat coleoptile segments. This enzyme was shown to increase cell wall extensibility as measured by a stretching method. In other studies, Masuda et al. (1970), compared the activity of the endo-b -1,3-glucanase and its activity on Avena coleoptile cells with those of the exoenzyme. They found that exoglucanase enhanced elongation and extensibility of the cell wall. But the effect was not additive to the effect of IAA. Furthermore, at least three hours of incubation with exoglucanase was required for enhancement of elongation. The endoglucanase showed no effect on cell wall elongation. Cell wall turnover and auxin effects thereon in pea stem tissue have been studied using pulse-chase wall labelling experiments. Considerable turnover of galactan occurs but this is not influenced by IAA (Labavitch & Ray, 1974).
The action of auxin on soluble xyloglucan can be assayed with relative ease and precision and has been positively demonstrated to be in progress within thirty minutes after exposure to IAA and probably within fifteen minutes, placing it amongst the most rapid metabolic effects of auxin. This auxin effect is blocked by metabolic inhibitors that are known to block elongation, but persists under complete osmotic inhibition of elongation by mannitol. It seems likely that it is involved in the action of the cell wall that leads to elongation in pea cells. Involvement of xyloglucan is understandable in terms of a recent model for cell wall structure, according to which xyloglucan serves to bind matrix polysaccharides to cellulose microfibrils (Bauer et al., 1973; see also chapter 1).
13.2.4—
Hydrogen-Ion Pump
Since the 1930'S it had been known that acidic media can stimulate elongation in auxin-sensitive tissue. Recent work showed that acidic buffers or CO2 solutions in the pH range from 3 to 4 induce a rate of coleoptile cell elongation as great as or greater than that obtainable with auxin (Rayle & Cleland, 1970; Hager et al., 1971; Evans et al., 1971). Acid-induced elongation starts almost immediately upon treatment rather than after the 10–15 minute latent period
characteristic of auxin action. Acid-induced elongation is not suppressible by metabolic inhibitors, such as cyanide, mercurials, and cycloheximide, or by lack of oxygen, all of which block auxin-induced growth, and appears to be a passive process independent of metabolism. In later work it was found that when the epidermis is removed to improve H+ entry into the tissue, the full acid-pH stimulation of elongation develops over the pH range from about 6.0 to about 5.0 (Rayle, 1973; Cleland, 1973).
Rayle et al., (1970b) found that cell wall skeletons of frozen-thawed, dead coleoptile tissue would elongate dramatically if treated with acidic media while being held under tension by an applied load. Rayle and Cleland (1972) concluded that acid pH- and IAA-induced elongation must occur by the same mechanism, because the two kinds of elongation had similar rate, similar temperature dependences, and a similar yield threshold. While suggestive, these similarities do not establish identity of biochemical mechanism.
Hager et al., (1971) also felt, on grounds that IAA-induced elongation could be suppressed by alkaline media, that IAA-induced growth involves the same biochemical mechanism as acid-induced elongation. In support of their proposed auxin-stimulated H+ pump they offered experiments showing rapid stimulation of coleoptile elongation by ATP, ITP, and GTP under anaerobic conditions, although these results may have represented merely the acid pH effect itself, since these compounds are strong acids and were applied in unbuffered, pH 5 media.
In recent work an auxin-induced release of H+ ions from coleoptiles stripped of their epidermis (Cleland, 1973; Rayle, 1973) and from other auxin-sensitive tissues (Marrè et al., 1973; Ilan, 1973) has been detected. This is measured simply as a gradual fall in pH of the medium bathing the tissue, upon treatment with IAA, from near-neutral pH to pH 5 or below, i.e., into the range that by itself induces elongation. Detectable H+ secretion by coleoptiles begins within 20 to 30 minutes after exposure to IAA and may be regarded as a 'rapid response'. To the extent so far studied, H+ secretion by coleoptiles has a specificity for auxin analogues and antagonists similar to the specificity seen in growth, and a similar dependence on metabolism. Secretion is sensitive to inhibitors and uncouplers of energy metabolism and, perhaps unexpectedly for a transport process but just like cell enlargement, H+ secretion is quickly inhibited by low concentrations of cycloheximide (Rayle, 1973; Cleland, 1973). Comparable results with pea stem segments have been reported by Marrè et al., (1973).
These findings, coupled with the observations that induction of elongation by auxin is prevented by sufficiently well-buffered media of pH 6 to 8 provided the cuticle is removed or rendered permeable by gentle abrasion (Rayle, 1973), constitute presumptive evidence that the auxin effect on cell enlargement is mediated by externally secreted H+ ions.
The simplicity and directness of the acid secretion theory of auxin action is appealing and the theory has quickly become popular. Various auxin effects on cation and anion transport noted previously were considered by their authors
to be consistent with the H+ secretion theory. H+ secretion could involve either a parallel flow of anions, or a counterflow of, or exchange with, cations as has been inferred for H+ pumping by beet cells (Poole, 1973). However, as yet no immediate dependence on external ions of either auxin-induced H+ secretion or growth has been found.
Cleland (1973) and Rayle (1973) observed that auxin-treated coleoptiles cease to release H+ when the medium reaches pH 5.0 or slightly below, suggesting that auxin-stimulated H+ secretion is inhibited by acid pH, like other H+ pumps (Poole, 1973), or alternatively that, at pH 5, back-diffusion of H+ into the cell offsets the action of the pump. This feature should allow extensionpromoting pH values in the cell wall to be reached rapidly under auxin treatment, while shutting down further acidification which would probably be injurious to the cell (Cleland, 1973).
Some objections to the H+ secretion theory of auxin action and weaknesses in the evidence for it should be mentioned. For example, it has not yet been shown that suppression of auxin elongation by neutral buffers is not due to inhibited uptake of auxin, or to failure of primary action (the auxin binding found by Hertel et al., (1972) had a pH optimum below pH 6.5).
The most important piece of evidence still needed for the acid secretion theory is that the pH in the tissue's free space falls to an extension-stimulating value at the time that rapid auxin-induced elongation commences, i.e., within 10–15 minutes after exposure to auxin, as against the hour or more that is required for an external bathing medium to reach pH 5.0 to 5.5 (Cleland, 1973; Rayle, 1973). At the very least it must be demonstrated that IAA induces H+ secretion as quickly as it induces rapid elongation. Existing data fail this test and show an increase in H+ efflux from coleoptiles beginning, at the earliest, 20 or 30 minutes after exposure to IAA (Cleland, 1973; Rayle, 1973). These authors feel that the discrepancy may be attributed to time lags imposed by the free space volume and the diffusion path length for H+ to reach the external medium. A worse timing discrepancy was seen with sunflower hypocotyl (Ilan, 1973; cf. Uhrström, 1969). A more sensitive method for measuring H+ efflux needs to be employed to resolve these discrepancies. Better still, the pH within the free space of the tissue should be measured directly during response to auxin treatment.
13.2.5—
Regulation of Genetic Material
After the discovery of messenger-RNA it was suggested that the regulation of gene action by auxin required the specific control of messenger-RNA synthesis. Using a series of inhibitors, Key (1964) and Key and Ingle (1964) noted that treatment of soybean hypocotyl with 5-fluorouracil (5-FU) inhibited total RNA synthesis by 80% whilst growth proceeded normally. Further studies on the fractionation of nucleic acid from soybean hypocotyls on a methylated albumin Kieselguhr column showed that 5-FU inhibited the incorporation of labelled
adenosine diphosphate into ribosomal-RNA but had little effect on the labelling of messenger-RNA. Additional experiments with soybeans showed that 5-FU did not affect the tenaciously bound RNA whose synthesis is promoted by 2,4-D.
Sen and his colleagues (Roychoudury & Sen, 1964; Roychoudury et al., 1965) showed that nuclei isolated from coconut milk responded to auxin by making more RNA in vitro. On the basis of these observations, it was proposed that auxin directly affected the nucleus and thereby regulated gene expression. It was thought that in the presence of the hormone a greater amount of DNA template was exposed and allowed to be transcribed into RNA. In an attempt to test this hypothesis, Cherry (1967) showed that nuclei isolated from peanut cotyledons did not respond in vitro to 2,4-D at physiological concentrations (10–8 to 10–6 M ). From many other experiments, Cherry demonstrated that only in approximately 1 out of 10 attempts could nuclei be shown to respond to the hormone in vitro by the synthesis of more RNA. In general, absolutely no effect of the auxin on the capacity of the isolated nuclei to produce RNA was found. On the other hand, it was found routinely that isolated nuclei from soybean seedlings pretreated with 2,4-D produced twice as much nucleic acid as did control nuclei. Furthermore, experiments of O'Brien et al., (1968) showed that chromatin isolated from soybean hypocotyls did not respond in vitro to 2,4-D. In all cases the tissues needed to be treated with the growth regulator for at least two hours before any effect on chromatin-directed RNA synthesis could be noted. It is to be noted, however, that in a few experiments an effect of the hormone could be observed within thirty minutes. Subsequently, a progressive increase in chromatin-directed RNA synthesis was noted as a function of time. From those experiments, it was suggested that the auxin-enhancement was a result either of a more active RNA polymerase or of gene derepression leading to an increased availability of DNA template. A third possibility was that both of these effects were involved. Subsequent experiments demonstrated that in the presence of E. coli RNA polymerase, chromatin from both control and 2,4-D treated plants allowed the synthesis of similar amounts of RNA at saturation with the polymerase. Even though the total E. coli RNA polymerase activity with chromatin from 2,4-D treated plants was slightly higher than that with control chromatin, it was concluded that the auxin primarily promoted the endogenous RNA polymerase, rather than the chromatin template availability.
When it became possible to solubilize RNA polymerase from chromatin (Hardin & Cherry, 1972) two important aspects were noted. First of all, it has not been possible to show that the addition of auxin in vitro to solubilized RNA polymerase, or to chromatin, increases the rate of RNA synthesis (Hardin et al., 1970). Secondly, of the many experiments that have been performed, it appears that treatment of sensitive plants with auxin leads to a greater production of RNA polymerase I (Hardin & Cherry, 1972; Hardin et al., 1972). This is the enzyme which is thought to be present in the nucleoli, as judged from comparisons with animal RNA polymerase, and is the enzyme thought to transcribe ribosomal into ribosomal-RNA. These data agree with the fact that large increases
in ribosomal-RNA and ribosomes are observed after auxin treatment. However, they are inconsistent with the idea that auxin increases the activity of the RNA polymerase enzyme which transcribes unique DNA sequences into messenger-RNA.
13.2.6—
Action on Membranes
A current, interesting view of auxin action is that the hormone rapidly effects its action at, or on, cellular membranes, possibly involving the regulation of the export of growth active materials across the plasma membrane into the cell wall space. Auxin action at the plasma membrane was hypothesized by Hertel and Flory (1968) and by Rayle et al. (1970) partly in the belief that auxin transport and auxin action on growth involve closely related, if not identical, interactions, presumably with some carrier site located in the plasma membrane. Evidence for auxin action at the plasma membrane comes from observed IAA effects on membrane potentials (Tanada, 1970). This effect, however, has not yet been adequately studied nor shown to be related to the elongation process. Another indication of auxin action at the plasma membrane is the induction of cell elongation by certain fungal toxins which are known to increase membrane permeability (Evans, 1973).
It is assumed that specific auxin receptor sites must be located in or on the plasma membrane if the hormone is to change the permeability or to change any physical property within the membrane. With this general assumption in mind, Lembi et al. (1971) began to look at the binding of labelled NPA (naphthylphthallamic acid) a powerful competitive inhibitor of polar auxin transport which was considered to be likely to bind to specific auxin binding sites. Using maize particle preparations, they tentatively showed that there was a greater amount of NPA binding to fractions rich in plasma membranes in comparison to all other fractions obtained. IAA did not compete with NPA for the sites which thus do not seem to be the actual carrier sites for polar transport of IAA. It is of course conceivable that the sites binding NPA are not the same sites which would normally bind the auxin, IAA.
Hertel et al. (1972) detected a specific binding of labelled IAA and naphthaleneacetic acid (NAA) to certain particles in maize homogenates. Specific binding was assayed as radioactivity retained by particles exposed to 2 to 10 × 10–7 M labelled auxin minus radioactivity retained when, in addition, 10–4M unlabelled IAA or NAA was added to compete with and displace labelled auxin from saturateable binding sites. The radioactivity which remained in the latter case then was termed unspecific binding. This difference-assay gave specific binding with binding constants in the rage of 10– ; to 10–5M . Specificity of the binding as an auxin phenomenon was indicated by the evidence that analogues of IAA or NAA which are active on growth, and are transportable, can displace labelled IAA or NAA from the binding sites, whereas chemically similar, but biologically inactive, molecules do not compete in the binding assay.
One defect of these data was that 2,4-D, a very active auxin, competed relatively weakly with IAA or NAA in the assay, nor was specific binding of labelled 2,4-D itself detected. Based on the evidence that auxin-specific binding sites are localized in plasma membranes, Hardin et al. (1972) proposed that the reaction of auxin, either the native IAA or the synthetic 2,4-D, with the plasma membrane would probably lead to conformational changes within the membrane, causing the release of a receptor molecule into the cytoplasm. This receptor molecule may or may not be the same binding agent with which the auxin interacts. Nevertheless, the receptor released from the plasma membrane could then travel through the cytoplasm and into the nucleus where we know auxin ultimately increases the activity of RNA polymerase.
From previously published information from animal systems, (Shyamala & Gorski, 1969; Jensen et al., 1968) a good analogy can be drawn with hormone interaction with receptor molecules which then migrate into the nucleus where nucleic acid synthesis is controlled. Following these lines of investigation, Hardin et al. (1972) isolated a plasma membrane-rich fraction in which the plasma membrane was thought to be 70% homogeneous. The addition of plasma membranes to soybean RNA polymerase caused an increase in RNA synthesis in vitro. Subsequently, it was shown that pretreatment of the membranes with 10–7M 2,4-D greatly increased RNA polymerase activity. Still further, when the plasma membranes were pretreated with 2,4-D followed by removal by centrifugation, the supernatant fraction contained the RNA polymerase stimulus. The chemical release of this factor appears to be specific for auxin, i.e., IAA and 2,4-D bring about this release, but a non-auxin such as 2,5-D is totally inactive.
In other studies an attempt was made to determine which of the multiple RNA polymerases were stimulated by the auxin-released factor. It was found that the addition of plasma membrane factor to a semi-purified RNA polymerase preparation enhanced the a -amanitin sensitive polymerase. By analogy with animal RNA-polymerase studies, these data imply that auxin increases the nucleoplasmic RNA poly merase, the enzyme which is thought to transcribe DNA into messenger-RNA. If this is true then it is sensible to believe that auxin, through the release of a receptor molecule, may regulate the transcription of unique DNA sequences through the control of a specific enzyme.
13.2.7—
Summary
Because of rapid auxin responses on plant cells, over the last few years a whole new field of research has begun which relates not only to the gene concept but also to rapid responses. Therefore, the new area of research has dealt with actions of auxins on receptors, membranes and other binding surfaces which could lead to a rapid growth response, as well as a continued and long response involving nucleic acid synthesis and protein synthesis.
The apparent binding of auxin to plasma membrane sites has stimulated interest in the concept that the hormone might bring about conformational
changes within the membrane structure. This in turn might lead to changes in permeability, causing an increased ion flux across the membrane. If a receptor molecule is located on the plasma membrane, a change in conformation of the membrane might bring about a release of the receptor. It has been proposed that this receptor moves into the nucleus of the target cell and once there, increases the activity of RNA polymerase. This, in turn, leads to the synthesis of messenger-RNA which codes for proteins and this brings about the net result of auxin increased growth.
In this particular section the effects of cyclic-AMP on auxin controlled growth have not been covered. When cyclic-AMP was first found to stimulate plant growth, considerable excitement was generated (Solomon & Mascarenhas, 1971). However, that excitement was short-lived, and it now appears that cyclic-AMP has little or no effect on plant cells (Ownby et al., 1975).
A number of enzymes have been shown to be affected by auxin. These include the hydrolases, particularly cellulase, and other enzymes associated with cell wall degradation as well as cell wall synthesis. Whilst these enzymes are affected by the auxins, it is very likely that this result is a secondary effect of the auxin and is not related to the primary mechanism of action. In essence then, it appears that auxins control a multitude of physiological and biochemical responses in plants. It is possible that the primary site of action is localized within the plasma membrane or some surface binding site of the cell. A change at this particular level, either in structure or function, could bring about the release of a factor which moves into the nucleus. As a secondary amplification, the hormone thus modulates the activity of RNA polymerase. This in turn leads to the synthesis
Figure 13.2
A hypothetical model of auxin action. The interaction of auxin with the
plasma membrane results in the release of a factor which moves through
the cytoplasm and into the nucleus. The factor controls the activity of RNA
polymerase II in the nuclei which leads to synthesis of mRNA. The new species
of mRNA translated in the cytoplasm into new proteins leads to enhanced cellular growth.
of RNA from parts of the genome which were either not transcribed at all, or just to a low level in the absence of the receptor. A change in DNA transcription in this manner would lead to the synthesis of new proteins which would bring about the change in the growth potential (Fig. 13.2).
13.3—
Gibberellin Action
13.3.1—
Introduction
Gibberellins may be defined as compounds having a gibbane skeleton and biological activity in stimulating cell division or cell elongation or both (various structures are given in Fig. 13.3). Gibberelins also may possess other biological
Figure 13.3
Structure of gibbane and the conversion
of GA4 to common gibberellins.
activities such as induction of nucleic acid and enzyme synthesis. Presently there are more than forty known gibberellins which have been isolated from green plants or the fungus Gibberella fujikuroi (Cherry, 1973). Since the late 1960's three to four new gibberellins have been identified each year. Thus, it seems that great progress is being made in the identification of naturally-occurring gibberellins; on the other hand the mechanism of action of gibberellins is still unknown. Plant physiologists are therefore confronted with a very wide array of gibberellins, several of which often occur in the same tissue, and the question arises as to their function within the cells.
Work beginning in the 1960's on the possible regulation of gene transcription by gibberellins was initially successful and achieved wide-spread acceptance. Later work raised many doubts, however, and at the present time, many workers in the field reject the concept that gibberellins regulate physiological processes through the control of gene transcription. Alternative mechanisms involving an action of gibberellins on lipids and membranes are now being proposed.
13.3.2—
Gibberellin Control of Enzyme Synthesis
The aleurone layer of cereal grain is composed of a relatively homogeneous population of non-dividing cells which respond as a target tissue to gibberellins in the concentration range of 10–7 to 10–10 M (Varner & Johri, 1968). It is, therefore, an ideal tissue for a study of the hormonal control of enzyme activity. Haberlandt in 1890 first discovered the secretion of starch liquifying enzymes by the aleurone layer in rye grain in response to some substance obtained from the germinating embryo. Since that early discovery, over the many years following, many workers have found embryo substances or factors and gibberellin-like materials which bring about an increase in activity of several hydrolase enzymes (Varner & Johri, 1968). Some of the enzymes which have been shown to increase in activity in the aleurone layers of barley grains in response to treatment with gibberellic acid (GA3 ) include the following: a -amylase, proteinase ribonuclease, b -glucanase, and pentosanases. A similar control of amylase activity by gibberellins has been observed in wheat and in wild oats. In barley aleurone layers GA-induced increase in a -amylase is prevented by the lack of oxygen, by cytochrome c inhibitors and inhibitors of RNA and protein synthesis. Varner and Johri (1968) have shown that gibberellin control of amylase appears to involve the control of nucleic acid and protein synthesis.
GA3 enhances the development of a -amylase activity in cereal grains. The enzyme is synthesized de novo and its synthesis blocked by cycloheximide and other inhibitors of protein synthesis. It is of interest to note that actinomycin D (an inhibitor of RNA synthesis) only inhibited the production of a -amylase when the tissue was exposed to the inhibitor early in the incubation process. If, however, the tissue was exposed to actinomycin D at some point after enzyme
synthesis had begun, little or no inhibition occurred. Thus, it was not certain whether the hormone actually controlled the synthesis of RNA at the level of transcription or controlled the translation of pre-existing messenger-RNA. Many attempts were initiated to answer this question; unfortunately, to date, a definitive decision has not been possible. The most likely conclusion is that the hormone does not control the synthesis of a -amylase at transcription (Chrispeels & Varner, 1967).
13.3.3—
The Effects on RNA Synthesis
Considerable indirect evidence has shown that GA3 influences the synthesis of RNA in many plant systems (Varner & Johri, 1968). If this RNA synthesis is a primary effect of the reaction of GA3 within a cell, it is then most likely that the primary action occurs in the nucleus. In many other plant tissues also, either an increase in protein and RNA synthesis or in the incorporation of labelled precursors into these macromolecules in the target tissues is evoked by hormones (Key, 1969).
It seems obvious that if GA interacts directly with the nucleus it would be advantageous to work with isolated nuclei or chromatin obtained from the nuclei. Therefore, attempts were made to work with nuclei isolated from light-grown dwarf peas (Varner & Johri, 1968). To determine if the RNA synthesized in the presence of GA3 was qualitatively different, the nuclei were incubated in the presence of one of the four nucleoside triphosphates labelled in the a position with 32 P and the radioactivity then determined in the 2',3' nucleotides. It was found that a significant difference existed in the relative distribution of radioactivity among the four nucleotides of the RNA synthesized in the presence and absence of gibberellic acid. This nearest-neighbour analysis indicated that RNA synthesized by GA-treated nuclei is in part qualitatively different from the RNA synthesized by untreated nuclei.
Fractionation of the labelled RNA on MAK columns showed differences also in the types of nucleic acids produced in the presence of GA3 . The results indicated that GA preferentially enhances the synthesis of a small fraction of RNA. However, fractionation of the labelled nucleic acids on sucrose gradients indicated that the in vitro synthesized RNA sediments in a peak corresponding approximately to 6–10s. These results indicate that the RNA synthesized by GA-treated nuclei has a higher average molecular weight than that synthesized by the control nuclei. Varner and Johri (1968) concluded that the enhanced RNA synthesis could be due (a) to an increase in template DNA availability, (b) to an increase in the RNA polymerase activity, or (c) to a combination of the above two possibilities. An enhanced activity of RNA polymerase could be due to activation or to protection against degradation of the enzyme. Another explanation might be that the hormone changes the permeability of the nuclear membrane and thereby makes the RNA precursors more available for use in transcription.
13.3.4—
Gibberellin Control of Enzyme Secretion
The gibberellin-enhanced synthesis of hydrolases is preceded by a proliferation of rough endoplasmic reticulum (Yomo & Varner, 1971). The control of rough endoplasmic reticulum seems more likely than the control of hydrolase synthesis and secretion as a candidate for the primary site of gibberellin action since it could more readily be a general response of plant tissues to gibberellins. Varner and his associates (Evins & Varner, 1971; Koehler & Varner, 1973) have therefore examined rough endoplasmic reticulum in detail. There is a gibberellinenhanced incorporation of labelled choline into the lipid-soluble material of the endoplasmic reticulum and of other cell fractions. An enhanced incorporation of 32 P into the phosphoryl-lipids of several cell fractions is also seen. This enhancement is first measurable about four hours after the addition of gibberellin and is proportional to the logarithm of the concentration of the hormone.
GA3 also enhances the activity of phosphorylcholine cytidylate-transferase and phosphorylcholine-glyceride transferase (Johnson & Kende, 1971). These responses are measurable as early as two hours after treatment. Gibberellin-evoked increases in these membrane-bound enzyme activities are prevented by the inhibitors, abscisic acid, actinomycin D and cycloheximide. These early effects on phosphoryl-lipid metabolism suggest that hydrolase synthesis occurs only on polysomes bound to newly synthesized endoplasmic reticulum. Thus the synthesis of hydrolase before the addition of GA3 may be limited not by messenger-RNA but by the availability of appropriate membranes for the attachment of polysomes which carry hydrolase-specific messenger-RNAs. An examination of the secretion of hydrolases is of interest because it appears that gibberellin is required for the secretion of b -glucanase which had been synthesized before hormone addition. Gibberellin-enhanced release of b -glucanase into the medium surrounding the aleurone cells is prevented by inhibitors and uncouplers of oxidative phosphorylation, by actinomycin D, 6-methylpurine, and by cycloheximide. This is in contrast to the secretion of a -amylase which requires phosphorylative energy transformation but is not affected by 6-methylpurine nor by cycloheximide. These differences in the secretion of b -glucanase and a -amylase may represent different modes of secretion for the two enzymes. Or, it may be that the cellular apparatus required for the secretion of both enzymes is made during the first few hours of gibberellin treatment.
13.3.5—
Effects of Gibberellins on Membranes
For the past few years Paleg and his associates have studied the effects of GA3 on various membranes and membrane properties (Wood & Paleg, 1974; Wood et al., 1974). Their studies were initially encouraged by the work of Bangham et al. (1965) who provided evidence that steroids have potent effects on model membranes composed only of lipids. Correlation between steroid activity in vivo
and in the model membrane systems was good. A suggestion was made that steroids produced their biological effect through a direct interaction with lipid, independent of polysaccharides, proteins or even active cell metabolism. The influence of GA3 on the permeability of lipid model systems was first described by Wood and Paleg (1972) who suggested that gibberellins might similarly exert their physiological action through an effect on lipids. Recently, Wood et al. (1974) have demonstrated complex formation between GA3 and phosphatidyl choline thus providing definitive evidence of a molecular association between the hormone and a common membrane component. In this study, complex formation was dependent on hormone concentration. Wood and Paleg (1974) also showed that the process of glucose diffusion from liposomes (i.e. microvesicles composed of a bimolecular lipid membrane surrounding aqueous contents) is not affected by the presence of GA3 . The same energy is required for a mole of glucose to permeate the lipid at each temperature in the presence or absence of the hormone. However the barrier to diffusion is perturbed in such a way that more glucose can pass per unit of time in the presence of GA3 . Furthermore, the temperature at which the lipid membrane undergoes a change in conformation (the transition point) is lowered by several degrees by hormone treatment.
By comparison with living systems, GA3 could, by its influence on lipid polarity, affect two main processes in vivo; (a) the permeability of the membranes to regulatory ions which act as limited substrates; and (b) the activity of membrane-bound enzymes such as ATPase which are activated by sodium, potassium, etc. Both of these effects could be brought about by the same mechanism—greater membrane fluidity. Since GA3 influences not only the transition point at which the liposome changes in structure but also the diffusion rate of glucose above this transition point, one or both of two mechanisms may explain its action. Either its presence directly reduces the viscosity of the central core of the membrane or, more likely, it perturbs the still-ordered polar regions of the liquid crystal phase leading to greater fluidity.
However, the temperature zone of greatest physiological interest is that where the untreated system is below the transition point while in the presence of hormone the system has switched to high temperature sensitivity. The relationship between biological activity of different gibberellins and their ability to influence the thermal transition and membrane permeability has not been explored extensively. However, the methyl ester of gibberellic acid, a derivative with only about 10–3 of the biological activity of unsubstituted GA, was found to be without effect on the temperature at which the transition of the membrane occurs. In this regard it is of interest that it also induces very little alteration of the nuclear magnetic resonance spectrum of lecithin, which indicates little complex formation between the hormone and the fatty acid. This new area of research may suggest that the mechanism of GA action is through the direct alteration of membrane structure and/or function.
13.3.6—
Summary
The gibberellins comprise a complex and large group of related compounds which control cell elongation and enzyme secretion. The mechanism by which the hormone turns on the synthesis of enzymes has been studied in fairly close detail. Initially, it was felt that the hormone regulated the synthesis of RNA at the level of transcription. Through the careful and dedicated work of Varner and his associates it appears reasonably clear that the hormone does not control the secretion of enzymes by controlling the synthesis of messenger-RNA. However, it is clear primarily from the work on amylase, that inhibitors of nucleic acid synthesis and protein synthesis disrupt the production of the enzyme. The recent experiments of Paleg and his associates bring a new dimension to the question of gibberellin action by their demonstration that GA3 affects the membrane permeability of fluidity. Thus, a new range of possible hormonal mechanisms can now be envisaged and explored.
13.4—
Action of Cytokinins
13.4.1—
Introduction
The term cytokinin has been accepted practically universally as a generic name for substances which promote cell division and exert other growth regulatory functions in the same manner as kinetin. The synthesis and testing of compounds for cytokinin activity began with the discovery of kinetin (6-furfurylaminopurine). Today there are probably at least one hundred known synthetic and native cytokinins (Fig. 13.4).
Structural requirements for a high order of cytokinin activity generally include an adenine molecule with the purine ring intact and with an N6 substituent of moderate size. One exception to the requirement for a modified purine exists, notably diphenylurea and its derivatives. Certain other substances which lack a true purine ring, such as 8-azakinetin, 6-benzylamino-8-azapurine and 6-(3-methyl-2-butenylamino)-8-azapurine are active, but each of them is less than 10% as active as its corresponding purine derivative. Substitution of either O or S for N in the N6 position of adenine in each case results in a more than 90% loss in growth promoting activity in the tobacco bio-assay.
Cytokinins play a role in practically all phases of plant development from cell division to the formation of flowers and fruits. They affect metabolism including the activities of enzymes and the biosynthesis of growth factors. They also influence the biogenesis of organelles and the flow of assimilates and nutrients within the plant. Cytokinins defer senescence and may protect the plant against adverse environments, such as water stress. These many diverse effects presumably stem from some primary anabolic function of the cytokinins that remains to be elucidated.
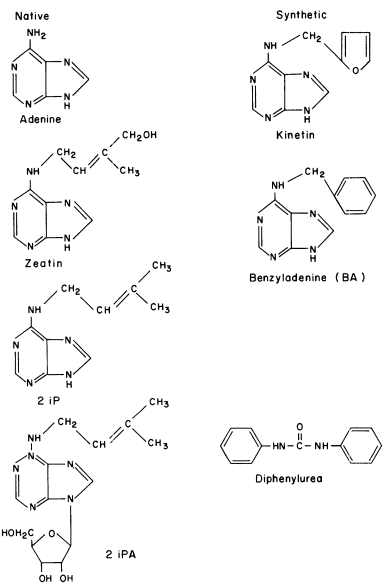
Figure 13.4
Structure of some common
native and synthetic cytokinins.
Cytokinins have been found in certain transfer-RNA (t-RNA) molecules from a large number of organisms including bacteria, yeast, plants and animals (see Cherry & Anderson, 1971). In sequence analysis the native cytokinin, isopentenyl adenine, always occurs adjacent (3' side) to the anticodon (Fig. 13.5). Furthermore, the location of the isopentenyl group is required for the
Figure 13.5
Diagram of the cloverleaf shape of seryl-transfer RNA.
The IPA residue is located adjacent to the anticodon.
specific tRNA to function efficiently in protein synthesis. Results involving bacteriophage infection of bacteria indicate that cytokinin in tRNA may have an important regulatory function in protein synthesis. In soybean seedlings (Anderson & Cherry, 1969) the application of a cytokinin (6-benzyladenine) results in increased levels of two species of leucyl-tRNAs. Although the relationship of cytokinins to tRNA may not be directly associated with the primary action of cytokinins on cellular activity, the alteration of specific tRNAs could control protein synthesis. Such a regulatory mechanism would be manifested in the control of growth and morphogenesis as the ultimate expression of the hormone.
13.4.2—
Actions of Cytokinins on Enzymes
Cytokinins have been shown to influence the activities of a number of specific enzymes. A most thoroughly studied case is the promotion by cytokinins of tyramine methylpherase activity in roots of germinating barley embryos. This appears to involve de novo synthesis although this conclusion is based solely on
the results of inhibitor experiments (Mann et al., 1963; Steinhart et al., 1964). Activities of four other enzymes tested were unaffected by kinetin treatment. The selective action on methylpherase is most likely related to the developmental stage of the seedling material and not to a specific affinity of cytokinin for the enzyme. Substances other than cytokinins (auxins and lysine) which also stimulated the appearance of tyramine methylpherase activity either were less effective or acted only during the early stages of germination. An analogous effect of kinetin in promoting synthesis of alkaloids in the roots of lupin has also been reported (see Skoog & Armstrong, 1970).
The induction of a -amylase activity in half-seeds without embryos, a well known effect of gibberellins in barley, is achieved in wheat also by treatment with cytokinins (Boothby & Wright, 1962). However, the effect of cytokinins on wheat is much less marked than that of gibberellin in the barley. In barley seeds, cytokinins cannot replace gibberellins, but do counteract an inhibition of gibberellin action by abscisic acid (Khan, 1969). The formation of amylase activity in excised hypocotyls of pea is stimulated by cytokinins but not by gibberellins or auxins (Chum, 1967). In germinating squash seeds cytokinins, but not gibberellins, at least partly substitute for the embryo in promoting the formation of isocitrate lyase and proteinase activity in the cotyledons.
The synthesis of deoxyisoflavones in soybean callus tissue is promoted by both cytokinins and auxins (Miller, 1969). The ease with which the compounds can be detected and the rapidity of the response suggests that this system might be useful as a rapid bioassay for cytokinins.
The rates of synthesis of ribulose bisphosphate carboxylase and NADP-dependent glyceraldehyde-phosphate dehydrogenase, studied in rice seedlings, appear to be under cytokinin control (Feierabend, 1969). Kinetin did not affect these enzymes until 96 hours after the beginning of germination. By contrast the activity of the cytoplasmic enzyme 6-phosphogluconic acid dehydrogenase was promoted by kinetin only during early stages of germination. It was concluded that cytokinins probably are not directly involved in gene derepression although the level of cytokinin may determine the extent to which the appropriate genes are expressed.
13.4.3—
Nucleic Acid Synthesis
Early studies with tobacco pith tissue showed that a proportion of the tetraploid cells present would be stimulated by kinetin treatments to enter mitosis and undergo cytokinesis without incorporation of thymidine. Thus the effect of kinetin on cell division was not always associated with DNA synthesis. In Lemna cultures 6-benzyl-aminopurine reverses the effect of abscisic acid in inhibiting growth and nucleic acid synthesis (van Overbeek et al., 1967). DNA synthesis in response to these growth substances appears to be more rapid than RNA synthesis. In axillary buds of tobacco which are normally inhibited by
the apex, treatment with 6-benzylaminopurine initiates DNA synthesis and growth (see Skoog & Armstrong, 1970).
Some inhibitors such as chloromycetin and 6-azauracil uncouple the two processes, preventing growth but not the cytokinin-induced DNA synthesis. Nevertheless, in view of the early work, it seems likely that cytokinin effects on DNA synthesis are indirect. In a detailed study of tuberization, Palmer and Smith (1969) have shown that, in excised stolons of potato, cytokinins inhibit stolon elongation and induce tuber formation. Other factors which inhibit elongation may also increase the inductive response in response to cytokinins, but they are not themselves inductive factors. Inhibitor studies suggest that cytokinin action is associated with protein synthesis or activation but not with DNA or messenger-RNA synthesis. Effects of cytokinin on RNA and protein synthesis and isolated cell organelles have been reported, but have not yet been analyzed in depth (Skoog & Armstrong, 1970). Nuclei preparations separated from coconut milk and incubated in the presence of kinetin showed an increased incorporation of labelled precursors into RNA and protein (Datta & Sen, 1965).
The effect of kinetin on RNA synthesis in an in vitro system containing purified chromatin and E. coli RNA polymerase has been studied by Matthysse and Phillips (1969). In the presence of a particular protein fraction, kinetin stimulated RNA synthesis. A similar effect of auxin was also mediated by a protein fraction. The effects of cytokinin in this system have not yet been studied in great detail. Fellenberg (1969) has observed that in vitro bindnig of kinetin to chromatin of pea epicotyls caused a slight decrease in the melting point of the nucleoprotein complex.
13.4.4—
Role of Cytokinins on Transfer RNA
One suggestion has been that the endogenous cytokinins somehow exert their activity by being incorporated into transfer-RNA (Skoog & Armstrong, 1969). For example, the synthetic cytokinin, benzyladenine, was reported to be incorporated into the tRNA of callus tissue (Fox & Chen, 1967). Richmond et al. (1970) specifically looked for incorporation of labelled benzyladenine into the RNA of senescing leaves, but found no evidence for incorporation. On the other hand, under similar conditions kinetin was incorporated into the RNA. Recently Walker et al. (1974) have shown that double-labelled benzyladenine is incorporated into tRNA at a frequency of one per 104 molecules.
Chen and Hall (1969) have shown that cytokinin-dependent tobacco callus tissue contains the enzyme system which attaches the D2 -isopentenyl group to the appropriate adenine residue in tRNA. In the case of tobacco callus tissue grown in the presence of radioactive benzyladenine, the tRNA still contained its complement of isopentenyl adenine derivatives even though the benzyladenine was incorporated into the tRNA. In general, various purine analogues can be incorporated into RNA in a rather non-specific way.
It is also quite possible that incorporation occurs into RNA fractions other
than tRNA, and unless the tRNA is rigorously characterized one cannot exclude the possibility of error arising by contamination with other RNA fractions. To establish whether the incorporation of N6 -substituted adenosine derivatives into tRNA has a biological significance it would be necessary to fractionate the tRNA and identify the incorporated material at a specific location in the tRNA sequence.
13.4.5—
Model for Cytokinin Action
Cytokinins are found in transfer-RNAs of animals, microorganisms and plants (see Cherry, 1973). Furthermore, in those cases where the nucleotide sequence is known the native cytokinin, IPA (isopentenyladenosine), or a modified form, is found adjacent to the anticodon. At the moment it appears that all IPA containing species of tRNA recognize codon triplets which begin with U (Skoog & Armstrong 1970). Thus, the anticodon loop of a cytokinin-containing RNA could be one of at least sixteen codon sequences of the following type: A— —IPA. By eliminating the nonsense terminator and the phenylalanine codons, it seems that IPA-containing tRNAs would be confined to less than ten codons (see Table 13.1).
|
It may be that IPA adjacent to the anticodon of these tRNA species plays some role in messenger-RNA-tRNA-ribosome recognition, and that these tRNA species are required for the translation of some messenger-RNAs into specific proteins. Based on these assumptions it is possible to speculate on the mechanism of action of cytokinins in plants. From the observation that cytokinins appear not to be incorporated into tRNA, as precursor units, but rather the isopentenyl group comes from mevalonic acid (Chen & Hall, 1969), it seems likely that cytokinins have no effect on the synthesis of tRNA containing IPA. Furthermore, a number of chemicals, including various isomers of IPA, kinetin, 6-benzyladenine and even diphenylurea, have cytokinin biological activities, and it appears that these cytokinins do not participate in tRNA synthesis. If this is true, then what is the mechanism of cytokinin action?
A model presented in Fig. 13.6 is based on the speculation that specific
Figure 13.6
A model of cytokinin action. Based on the fact that tRNA specific nucleases
are known, it is suggested that free cytokinins protect cytokinin-containing
tRNAs by inhibiting nuclease activity, specific as well as general.
nucleases break the primary structure of the IPA containing tRNA (Cherry & Anderson, 1971). The IPA would provide a unique attachment for the enzyme to bind to and subsequently to break the phosphodiester bonds. The action of this enzyme would destroy the function of the tRNA and yield free cytokinin. Cytokinins including 6-benzyladenine and diphenylurea added exogenously to plants or tissue cultures would defer senescence by essentially protecting the cytokinin-containing tRNA species. This protection would be mediated by the exogenously added cytokinin binding to the nuclease and competitively inhibiting its action. Alternatively, the cytokinin could bind to other agents which in turn prevent an increase in nuclease activity which would destroy the cytokinin-containing tRNA. Therefore, tissue treated with cytokinin would retain its IPA-containing tRNA species and continue to synthesize essential proteins as long as sufficient free cytokinin was present within the tissue.
Preliminary unpublished work of Locy (1974) provides suggestive evidence of a tRNA-specific nuclease. We have found that chloroplasts of soybeans contain a nuclease which specifically degrades one of the three tyrosyl-tRNAs. At the moment we do not have any evidence to suggest that free cytokinin controls the activity of this tRNA nuclease. In regards to this model it is to be emphasized that it is hypothetical and is presented as a working base upon which to investigate further the mechanism of action of cytokinins at the molecular level.
13.4.6—
Summary
Cytokinins have been shown to increase the activities of a number of enzymes. These increases in activity do not appear to be a direct effect of the cytokinin
but rather to be an indirect effect of some kind. Cytokinins increase the amount of DNA produced in a number of tissues. Furthermore, cytokinins increase the rate of RNA synthesis in ageing tissue. However, a number of studies have pointed out that the cytokinin effect on nucleic acid synthesis most likely is not due to a direct effect on the derepression of the DNA within the genome.
Over the past few years there has been a great deal of excitement generated relating to the fact that the native cytokinin is found in some transfer-RNA isoaccepting species. For a few years data was presented which showed that cytokinins, particularly the synthetic cytokinin, 6-benzyladenine, was incorported directly into transfer-RNA. That excitement, however, appears to be shortlived since other studies have shown that the incorporation of the labelled synthetic cytokinins is not into tRNA species but into heavier forms of RNA. Therefore, it appears quite clear that cytokinins are likely not incorporated into tRNA directly. A model is presented (Fig. 13.6) which suggests that the free cytokinins act as a protective agent within the target cells of plants to protect the transfer-RNA molecules which contain the native cytokinin. It is suggested that there are specific nucleases which degrade only certain species of transfer RNA. The model implies that free cytokinin inhibits the nucleases through product inhibition, or possibly through some other mechanism which prevents the enzyme from attacking the tRNAs which contain the cytokinin. At present there is only a small amount of evidence to substantiate this model.
13.5—
Actions of Ethylene
13.5.1—
Introduction
Ethylene is the simplest hormone which regulates plant growth (Fig. 13.7). It is a natural constituent of plants and affects a wide array of physiological processes. For many years, however, investigations of ethylene physiology were concerned primarily with fruit ripening. With the advent of gas chromatography many experimenters have begun research on the biochemical and physiological action of ethylene. The production of ethylene by germinating seeds and seedlings suggests that the hormone may be involved in the normal regulation of growth and development. Evidence for such a proposal includes the fact that low concentrations of applied ethylene blocks photo-induced apical bud expansion and hook opening in etiolated pea seedlings. It was shown that apical tissues of
Figure 13.7
Structure of ethylene.
etiolated seedlings are the major site of ethylene production. Light has been found to decrease the tendency of pea stem segments to produce ethylene in response to high concentration of auxin.
On the other hand, red light appears to stimulate ethylene production in dormancy. Another important role of endogenous ethylene in etiolated seedlings is the regulation of radial expansion of the pea epicotyl in a region below the apical hook. The exposure of the plant to red light or to CO2 inhibits an ethylene-mediated increase in the diameter of the epicotyl.
Growth, flowering, abscission and fruit ripening all are affected by ethylene. It is currently popular to speculate that the mode of action of ethylene involves a mechanism which regulates some aspect of the transcription of DNA or the translation of RNA. Studies of the effects of ethylene on abscission and growth indicate sizeable changes in RNA and protein contents. There is indeed evidence of changes in the activities of catalase, peroxidase, and other hydrolases. Furthermore a study showed that exposure of soybean plants to ethylene significantly altered RNA polymerase activity associated with chromatin. As judged by nearest neighbour analysis the RNA produced from ethylene treated plants has a different base composition than that from the control. Even though ethylene at very low concentrations affects a wide array of physiological and biochemical processes in plants, much more work is required to know the mechanism of action.
13.5.2—
Effect of Ethylene on Enzymes
A number of investigators have examined the possibility that ethylene has a direct effect on enzyme activity (Abeles, 1973). However, investigations with b -glucosidase, emulsin, a -amylase, invertase, peroxidase, and adenosine triphosphatase have shown that ethylene has no effect on these enzymes. Results reported by Abeles (1973) have similarly shown that ethylene has no effect on cellulase and carbonic anhydrase. Carbonic anhydrase was chosen because it appeared to be a likely candidate to show a positive response. Carbonic anhydrase contains zinc and has the ability to combine with CO2 , both features which suggest potential sensitivities for ethylene action. Nelson (1939) reported that ethylene increased the activity of trypsin. However, this effect was thought to be due to the removal of O2 since H2 had the same effect.
13.5.3—
Actions on Membranes
Since ethylene is more soluble in oil than in water and since membranes contain large amounts of lipid material, a number of investigators have tested the idea that ethylene affects some aspect of membrane permeability. However, proponents of this idea have failed to note that CO, an ethylene analogue, does not share the same lipid solubilities as ethylene but nevertheless has similar physiological activities. The idea that ethylene has a disruptive effect on the membrane
causing a change in permeability and an alteration in compartmentalization does not appear valid. Ripening fruits exhibit obvious changes in terms of permeability and retention of soluble components and it seems natural to suggest that ethylene leads to changes in membrane permeability which in turn causes softening and increased respiration. However, current evidence suggests that a change in membrane properties is an effect of ripening rather than a cause.
A similar situation exists in flowers. Nichols (1968) pointed out that solute leakage increased from carnations during senescence. Senescence and leakage were promoted by ethylene and reversed by CO2 . Ethylene has no influence on membrane permeability of potato, pea, avocado, banana, and bean. However, on the other hand, von Guttenberg and Beythien (1951) reported that ethylene increased the rate of deplasmolysis of
13.5.4—
Enzyme Induction by Ethylene
Regeimbal and Harvey (1927) were the first to report that ethylene-treated tissues contained higher levels of particular enzymes than the control. They found that ethylene increased the level of protease and invertase extracted from pineapple fruits. Since that time, reports (see Abeles, 1973) have appeared which show effects of ethylene on a number of enzymes. This list includes acid phosphatase, ATPase, a -amylase, cellulase, chitinase, chlorophyllase, cinnamic acid 4-hydroxylase, cytochrome c reductase, diaphorase, b -1,3-glucanase, invertase, malic enzyme, pectin esterase, peroxidase, phenylalanine ammonia-lyase, polygalacturonase, polyphenyloxidase, protease and pyruvic carboxylase.
Enzyme induction does not always depend entirely on the action of ethylene. In some cases, cutting the tissue causes an increase in enzyme activity. The function of ethylene is to reduce the lag time or increase the rate of increase in enzyme activity. Examples of enzymes whose increased activity does not strictly depend on ethylene action include b -1,3-glucanase, malic enzyme, phenylalanine
ammonia-lyase and peroxidase (Abeles, 1973). Excising tissue can cause wound ethylene production and it is possible that this source of ethylene plays some role in enzyme induction in tissue slices. On the other hand, enzyme induction during abscission was dependent on ethylene and for a reasonable period of time no cellulase synthesis or abscission occurred following excision unless ethylene was added to the gaseous phase.
13.5.5—
Effects of Ethylene on RNA Synthesis
The first report of ethylene effects on RNA synthesis was by Turkova et al. (1965) who reported an increase in RNA synthesis during epinasty of tomato leaves. Whether or not RNA synthesis was required for epinasty was not shown, although the idea is intriguing. Studies with actinomycin D indicated that RNA synthesis occurred during abscission and was required for the process to occur (Abeles & Holm, 1966). Support for this interpretation stems from the work of a number of investigators (see Abeles, 1973). It is known that the increase in RNA synthesis precedes that in protein synthesis and is localized at or near the separation layer. The increase in RNA occurred in all fractions including messenger-RNA, ribosomal-RNA and transfer-RNA. Differential extraction of the nucleic acids indicated that the ethylene-stimulated fraction was confined to that portion of RNA extracted by sodium lauryl sulphate with the increase found in ribosomal- and messenger-RNAs. The inhibitor, 5-fluorouracil, which blocks 50% of the ethylene-enhanced incorporation of 32 P into RNA did not inhibit abscission. The greatest inhibition occurred in transfer-RNA and ribosomal-RNA which indicates that the synthesis of all fractions of RNA is not required for abscission. Presumably as long as messenger-RNA was being synthesized, enough ribosomal- and transfer-RNAs were already available within the cell to permit abscission to take place. When all RNA synthesis was blocked by actinomycin D, abscission stopped (Holm & Abeles, 1967). Ethylene has also been found to promote RNA synthesis in preclimacteric fruit. Marei and Romani (1971) reported, as in the case of abscission, that the synthesis of all classes of RNA in fig fruit was stimulated. Holm et al. (1971) found that ethylene increased RNA synthesis in apples and that the increase in RNA was followed by an increase in protein synthesis. However, Sacher and Salminen (1969) reported that they failed to find an increase in RNA synthesis when preclimacteric bananas or avocados were treated with ethylene.
13.5.6—
Effects of Ethylene on Chromatin Activity
Holm et al. (1970) have reported that ethylene inhibited the growth of the apical part of soybean seedlings and caused an increase in the elongating and basal portions of the stem. At the same time, RNA levels in the apical zone were reduced while the levels were enhanced in the elongating and basal regions. Chromatin from the various parts of the seedling were studied to determine if the
capacity for RNA synthesis was also modified. They found that the activity in the apex was reduced while the activity in the elongating and basal regions were promoted. The rate of the response was rapid since the increase in chromatin activity was apparent after 3 hours. Nearest-neighbour analysis of the RNA synthesized demonstrated that there was a qualitative difference between RNA synthesized from chromatins of normal and ethylene-treated tissue. It was concluded that ethylene can regulate RNA synthesis as manifested by a change in quantity and kind of RNA.
13.5.7—
Effects of Ethylene on DNA Metabolism
Plant growth is either promoted, inhibited or unaffected by ethylene depending upon the tissue involved. Examples of growth promotion are swelling, epinasty, hook closure, seedling elongation and seed germination (Abeles, 1973). Bud break is probably a special case. Here no growth takes place as long as ethylene is present. However, after the gas is removed growth of the buds ensues. Growth inhibition is seen as arrested development of buds, leaves or apical meristems. Mature tissue, such as stems and leaves. do not undergo any change of size or weight, although premature senescence usually occurs. Since growth, or the lack of it, may be associated with cytokinesis, it is of interest to learn that DNA synthesis is controlled by ethylene. Holm and Abeles (1967) reported that DNA synthesis or DNA content in bean leaf tissue was not affected by seven hours exposure to ethylene, although abscission was promoted. Later they found that in soybean seedlings treated with ethylene the synthesis of DNA was inhibited in the apex where growth was inhibited but was promoted in the subapical part where swelling took place. Burg et al. (1971) found a similar situation in pea seedlings in which inhibition of cell division, measured as a reduction in metaphase figures, occurred within two hours after ethylene was added. However, it is not clear whether the change in DNA synthesis was the cause or the result of inhibited growth. Ethylene slows the growth of pea seedlings very quickly; for example, Warner and Leopold (1971) found growth slowed six minutes after ethylene was introduced into the gas phase and returned to normal sixteen minutes after the ethylene was removed. Kinetic studies on changes in DNA or other postulated sites of action are thus required to establish the relationship between cause and effect. Burg et al. (1971) have suggested that ethylene regulates DNA synthesis by some action on microtubule structure essential for spindle fibre formation during mitosis. If the action of ethylene was directed toward microtubules, this might also explain the reorientation of cellulose microfibril deposition that occurs in swelling.
13.5.8—
Summary
Even though little is known about the binding site(s) of ethylene, in a few cases such as ripening, abscission, swelling and senescence there is reason to believe
that the combination of ethylene with a site results in the regulation of protein or enzyme synthesis which in turn accounts for the observed responses. In these cases RNA synthesis, presumably messenger-RNA, with the accompanying support of soluble- and ribosomal-RNA, is an essential or early step suggesting regulation of gene action. However, in other cases, especially those associated with the physiology of excised tissue, control is probably not exerted at the level of RNA or protein synthesis. In the case of the inhibition of elongation, the action is apparently directed toward blockage of DNA metabolism. The site of action of ethylene in epinasty, root initiation, intumescence formation and floral initiation is even more poorly understood. The only valid conclusion appears to be that a number of essential features of plant growth and development are susceptible to ethylene action. In the final analysis it is concluded that there may be as many mechanisms of ethylene action as there are modes of ethylene operation.
13.6—
Actions of Abscisic Acid
13.6.1—
Introduction
Abscisic acid (ABA) is a plant hormone which now ranks in importanance with the auxins, gibberellins, cytokinins, and ethylene. Interest in the physiology and chemistry of ABA has grown greatly since the structure was established in 1965. During the 1950's and early 1960's a number of laboratories were engaged in research on growth-inhibiting substances. ABA was first isolated from cotton plants and was named abscisin II by a team from Addicott's laboratory (Ohkuma et al., 1963). During the same year Wareing's research team (Eagles & Wareing, 1963) isolated an active substance from Acer leaves which they named dormin. Abscisin II and dormin are the same substance which is now called abscisic
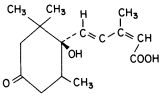
Figure 13.8
Structure of abscisic acid.
acid (Fig. 13.8). Plant tissues of all ages appear to synthesize and inactivate ABA. The number of various plant responses affected by ABA is very large. Generally, the physiological processes are related to senescence or abscission and growth retardation or inhibition. ABA appears to act as an abscission accelerating hormone in many fruits and leaves. Furthermore, it also tends to induce dormancy in some woody plants. ABA has been shown to move from the leaves to the apical bud to bring about a dormant condition. In potato, the
levels of inhibitors, including ABA, decrease during the quiescent period prior to renewed growth. ABA in extremely low concentration, moreover, prolongs the dormancy of excised potato buds.
Recently, ABA has been shown to be involved in the responses of many plants to stress conditions. As the ABA concentration increases in the leaves the stomata close. In this way, it appears that ABA is directly involved in the opening and closing of the stomata, thus regulating the rate of transpiration. Through this mechanism, ABA appears to protect the plant through conditions of water stress or drought.
At present the mechanism of action of ABA is not clearly understood. However, the available evidence indicates that ABA affects transcription as shown by reduced activity of chromatin-associated RNA polymerases. In other cases the mechanism of action of ABA appears to involve regulation of the translation of long-lived messenger-RNA, whilst its effects on stomata probably involve the regulation of membrane permeability.
13.6.2—
Role of ABA in Dormancy
One of the lines of research which led to the isolation of ABA from sycamore leaves was a change in the growth inhibitory activities in extracts of tree seedlings grown on long- or short-days. An increase in content of the growth inhibitory material in leaves was noted during the late summer and early autumn (Phillips & Wareing, 1958). Furthermore, the idea that bud dormancy in potato tubers may be caused by growth inhibiting substances had been suggested by Hemberg (1949). The original correlation of inhibitory material in leaves grown under short- and long-days and the induction of dormancy were based on measurements obtained by bioassay techniques. Recently, Lenton et al. (1973) have attempted to repeat these determinations using gas chromatography to compare the amounts of ABA present. They found that transferring birch, red maple or sycamore plants to short-days had no effect on ABA content. The importance of a balance between growth promoters and growth inhibitors has been stressed frequently. Tinklin and Schwabe (1970) have found considerably more inhibitor in bud scales than in the bud axis of blackcurrant. In these experiments the ABA concentrations were correlated with the degree of dormancy and the levels were reduced by treatments which encouraged bud break.
The most convincing evidence suggesting that ABA induces dormancy is the production of turions in Lemna polyrrhiza. When ABA is added to the medium under conditions that allow continuous growth, it not only inhibits growth but causes production of the dense, dormant, fronds known as turions (Stewart, 1969).
ABA is a potent inhibitor of seed germination, and its presence as a major growth inhibitor in dormant seeds of many species has cast it in the role of the maintainer of seed dormancy. ABA has been isolated from seeds of many genera of higher plants, and seeds of an equally large number of species have
been prevented from germination by soaking in ABA solutions. When seeds and fruit parts are separated, it is usually found that the concentration of ABA in the fruit is about 5 to 10 times greater than in the seed (Milborrow, 1974).
Lipe and Crane (1966) found that ABA in peach seeds decreased during stratification. Obviously, the part played by endogenous growth promoting compounds and the balance between them and ABA during the breakage of dormancy needs to be explored in more detail. Certainly, the balance between the two kinds of regulators is important. ABA is not an irreversible inhibitor since one of its most striking features is the facility with which it can be leached out of treated seeds thereby allowing the resumption of growth.
13.6.3—
Effects on Abscission
In the last few years several hundred experiments have been carried out in which plants, and parts of plants, of many genera have been treated in a variety of ways with ABA. It is quite unusual for any report to show clearly that the hormone controls abscission of leaves, which leads to the conclusion that exogenously applied ABA has little effect on leaf abscission. Nevertheless, ABA was first isolated by following its abscission-accelerating activity in petiolar stumps of cotton explants. The growth inhibitory action of this factor was believed to be responsible for the premature abscission of immature young lupin fruits (Cornforth et al., 1966). Consequently, the abscission-accelerating effect of ABA is well documented and extensively discussed (Milborrow, 1974).
Many of the experiments reporting leaf abscission have been carried out on tree crops using extremely high concentrations of ABA and often near the end of the growth season in attempts to cause abscission of fruits. The observed stimulation of leaf abscission may be an indirect effect of non-physiological high concentrations which stimulate ethylene production (Edgerton, 1971). For example, Cooper and Henry (1968) treated orange trees with sprays of 500 µg ml–l of ABA in summer and winter. The summer treatment caused leaves to develop colour and fall, but the winter treatments had no such effects. Olive trees were found to suffer some leaf abscission in one experiment but no effect was observed in another.
It appears, therefore, that ABA is not closely involved in the regulation of leaf abscission. The test system in which it has a stimulatory action consists of isolated petiolar explants containing presumptive abscission zones and maturing leaves nearing the end of their life. Even this tissue requires application of abnormally high concentrations of ABA to manifest an effect.
The role of ABA on fruit abscission is more certain. The early work of Van Steveninck (1959) showed that the inhibitor now known as ABA was intimately involved in the abortion of the young immature fruit of lupin. Application of ABA to mature peach, olive, apple and citrus fruits accelerated abscission and the effect of ABA was also marked on young grape flowers and berries (see Milborrow, 1966).
13.6.4—
Effects of ABA on Wilting
Wright (1969) found that when cut shoots were wilted there was an increase in the concentration of the so-called b -inhibitor. He went on to identify this inhibitor as ABA and defined the conditions under which the increase occurred. A water loss of about 10% in total fresh weight caused approximately 40-fold increase in ABA content while further water loss had no additional effect. Wright and Hiron (1969) have reported that other stress conditions, such as waterlogging, caused a similar rise in ABA, but they point out that such treatments cause wilting by reducing the efficiency of water uptake. The surprising feature of the increase in ABA is the rapidity with which it occurs. The content in turgid bean leaves has been calculated from the results of Wright and Hiron (1969). They show that ABA content increases from a normal level of 6 µg kg–1 fresh weight to 7 µg kg–1 within 7 minutes after blowing a dry and warm air stream across the plants. This level increases to 33 µg kg–1 within 25 minutes and 68 µg kg–1 within 45 minutes. In other experiments they show that in wheat leaves the ABA content increased from 23 to 171 µg kg–1 fresh weight within 4 hours of wilting. The ABA content of bean leaves remained at 67 µg kg–1 while they were kept waterlogged for 5 days. The implications of these data have yet to be explored in detail, but the observations offers a feasible explanation of the reduced growth of crops suffered during drought.
The extra ABA is probably formed by synthesis rather than by release from a precursor or conjugate because much more labelled mevalonate was incorporated into ABA by a sample of leaves that had been fed and then wilted than similar leaves which were kept moist during the entire experiment. Furthermore, the presence of 40 times the amount of a precursor or conjugate probably would have been detected by extraction and bioassay techniques. Many types of experiments have shown dramatic increases in ABA content on wilting in french beans, brussel sprouts, sugar cane, wheat, avocado spinach, cotton, peas and tomatoes (see Milborrow, 1974).
Perhaps the best information supporting a direct involvement in the direct closure of the stomata is afforded by a wilty tomato mutant produced by X-irradiation (Imber & Tal, 1970). The shoots of this plant contain one-tenth the amount of ABA contained in the normal variety. Applications of 0.1 to 10 µg of ABA caused a rapid and progressive reduction in transpiration rate of leaves and leaf discs. With 10 µg ABA ml–1 the stomata closed in darkness.
13.6.5—
Affects of ABA on Enzyme Activities
The ability of plant hormones to affect directly the activity of enzymes has not been extensively investigated. When an effect has been reported it is smaller than would be expected if the hormone specifically regulates at that site. Hemberg (1967) reported that ABA can inhibit a -amylase in vitro, but as the enzyme used was extracted from fungi the significance as related to higher
plants requires additional investigation. ABA has been shown to complex with fungal a -amylase and thereby change its physical properties. Saunders and Poulson (1968) found a slight stimulation of invertase activity at 10–7 M ABA and slight inhibition at 5 × 10–7 M and above. Again the significance is difficult to assess since a fungal enzyme was used.
13.6.6—
Effects of ABA on Nucleic Acid Synthesis
The first observations of the effects of ABA on nucleic acids were made by Van Overbeek et al., (1967) using Lemna. The incorporation of radioactive phosphate (32 P) into nucleic acid fractions was inhibited by ABA but reversed by benzyladenine. The primary site of action of ABA cannot be deduced with certainty because of the time scale and the complexity of the responses. Subsequent work has indicated that DNA synthesis is almost certainly not the primary target of ABA (Villiers, 1968). This has been clearly shown to be the case in dry wheat embryos by Chen and Osborne (1970) who found that protein synthesis commenced from imbibition and was inhibited at 6 hours by ABA, whereas RNA synthesis, as measured by the incorporation of 3 H-uridine, was not measurable until 12 hours. In the same experiments the incorporation of 3 H-thymidine into DNA was measurable after 24 hours. Experiments by Pearson and Wareing (1969) have shown that chromatin-directed RNA polymerase activity was slightly inhibited by 0.26 m g ml–l of ABA when added to the grinding medium. However, when ABA was added to the purified chromatin little or no effect was noted. Bex (1972) also reports that ABA has no effect on the binding between nucleohistones and DNA as measured by their melting point measurements. Schwartz (1971) has shown that ABA can alter the balance of alcohol dehydrogenase in maize, but the inhibitor was added during the growth of the cells. Thus, while it appears that ABA has small and random effects on nucleic acid biosynthesis in a number of plants, it is reasonably clear that the hormone has little or no direct effect on the synthesis of nucleic acids at the level of the genome.
13.6.7—
The Involvement of Abscisic Acid in Messenger-RNA Translation
Ihle and Dure (1972) investigated the appearance of various enzyme activities during the germination of cotton seeds and embryos. They showed that protease and iso-citrate lyase appear to be synthesized de novo during germination utilizing messenger-RNA which had been transcribed much earlier when only about 60% completion of embryogenesis had been reached. The translation of these messenger-RNAs during the last 40% of embryogenesis was apparently prohibited by the presence of ABA diffusing into the embryo from the ovule wall. The mode of action of ABA in maintaining translation inhibition appears to
involve RNA synthesis as judged by the fact that the ABA inhibition was inhibited by the presence of actinomycin D. Translation of the required messenger-RNAs for the germination enzymes may be induced prematurely by simply dissecting the ovules from the plant, suggesting that the breakage of the connection between the ovule and the mother plant may be required for the in vivo induction of translation. This hypothesis is further substantiated by the observation that, in vivo, the connection between the ovule and the placenta is normally severed at 60% completion of embryo-genesis.
The results coming from Dure's laboratory are the only ones at present which show that ABA may have a direct effect on the translation of messenger-RNA pre-existing in the embryo tissue.
13.6.8—
Summary
Even though ABA was first found in cotton plants and dormant sycamore leaves, it seems reasonably clear that this hormone has little or no effect on either the abscission of leaves or the dormancy of buds. It appears that the most likely physiological action of ABA on plants is to control the abscission zone in the fruit pedicel and thereby allow abscission to take place and cause fruit drop. Secondly, ABA very definitely appears to be involved in stomatal closure in response to stress conditions, particularly drought. In terms of a more biochemical approach it appears that ABA has little effect on various enzyme activities. Those enzymes studied show only small enhancement, or in some cases, inhibition, by ABA. Generally it has been shown that ABA inhibits the synthesis of various nucleic acids.
Again, one must be drawn to the conclusion that ABA has little effect directly on nucleic acid synthesis at the genome level. The most excitingp aproach to abscisic acid action appears to be that found in the cotton embryo where it has been shown to inhibit the translation of pre-existing messenger-RNAs found in the developing embryos. Even this response probably involves more complex processes than a direct inhibitory effect, since actinomycin D inhibits the ABA-induced inhibition. These data possibly imply that a suppressor molecule has to be formed to bring about effects.
Further Reading
Abeles F.B. (1973). Ethylene in Plant Biology. Academic Press, N.Y. 302 pp.
Cherry J.H. (1973). Molecular Biology of Plants, A Text-Manual. Columbia University Press. N.Y. 204 pp.
Hall R.H. (1973). Cytokinins as a probe of developmental processes. Ann. Rev. Plant Physiol.24, 415–44.
Jones R.L. (1973). Gibberellins: their physiological role. Ann. Rev. Plant Physiol.24, 571–98.
Key J.L. (1969). Hormones and nucleic acid metabolism. Ann. Rev. Plant Physiol.20, 449–74.
Milborrow B.V. (1974). The chemistry and physiology of abscisic acid. Ann. Rev. Plant Physiol. 25, 259–307.
Runeckles V.C., Sondheimer E. & Walton D.C. (eds) (1974). The Chemistry and Biochemistry of Plant Hormones. Vol. 7. Recent Advances in Phytochemistry. Academic Press. N.Y. 178 pp.
Skoog F. & Armstrong D.J. (1970). Cytokinins. Ann. Rev. Plant Physiol.24, 359–84.
Varner J.E. & Johri M.M. (1968). Hormonal control of enzyme synthesis. In Biochemistry and Physiology of Plant Growth Substances (eds. F. Wrightman & G. Setterfield). The Runge Press Ltd. Ottawa. pp. 793–814.
Chapter 14—
Phytochrome Action
14.1—
Introduction
Light, in addition to providing the energy which drives photosynthesis, also acts as a regulatory environmental stimulus. Many light-controlled plant responses are now believed to be mediated by the photoreceptor, phytochrome. This pigment has a regulatory role in all phases of plant growth and development (photomorphogenesis) and is apparently ubiquitous in all taxonomic groups of eukaryotic plants with the exception of fungi.
The central question of the molecular mechanism of phytochrome action is, however, yet to be resolved. Approaches to this problem fall into two broad categories: (a) Examination of the physical and chemical properties, localization and behaviour of the phytochrome molecule itself; (b) Investigations of the diverse array of phytochrome-mediated plant responses demonstrable at almost any level from the molecular to the gross morphogenetic. The principal findings and concepts that have emerged from such studies are summarized next. Some of the evidence that has led to these concepts is examined in subsequent sections.
14.2—
Phytochrome Dogma
Phytochrome is a blue-green biliprotein. Two principal forms of the molecule are readily distinguishable on the basis of their absorption spectra (Fig. 14.1)
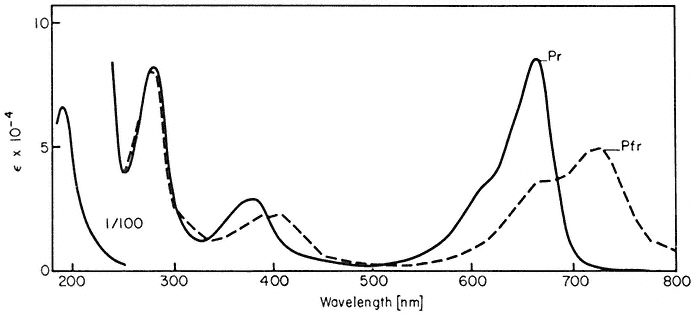
Figure 14.1
Absorption spectra of purified oat phytochrome following saturating
irradiations with red and far-red light (after Anderson et al., 1970).
and biological activity. The Pr form (= red-absorbing, lMax = 660 nm) is considered biologically inactive, whereas the Pfr form ( = far-red-absorbing, lMax = 730 nm) is considered to be the active species i.e. capable of inducing a biological response.
The two forms are reversibly interconvertible by red and far-red light ('phototransformation'). In addition, whereas the Pr form is stable in the dark, Pfr can revert thermally to Pr ('dark reversion') or undergo an irreversible loss of photoactivity ('destruction'). These properties are schematically represented thus:
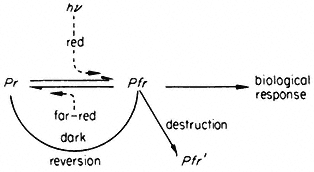
Red light absorbed by Pr converts the molecule to the Pfr form which in turn induces a measurable biological response. Far-red light, dark reversion and destruction provide alternate pathways for the removal of Pfr and thereby the potential for reversing induced responses. These properties endow the molecule with the capacity to function as a reversible biological switch, monitoring the environment for the presence, absence, intensity and spectral quality of photomorphogenically active light.
In molecular terms, phytochrome is considered to mediate the transmission of the light stimulus to the' cell via photoconversion to an active effector. Two major theories on how the cellular response system recognizes the effector have been advanced. One proposes phytochrome interaction with the genome; the other, interaction with cellular membranes.
14.3—
The Phytochrome Molecule
14.3.1—
Molecular Properties
The purified phytochrome molecule is a water-soluble chromoprotein containing less than 4% carbohydrate (Briggs & Rice, 1972). The native monomer is a polypeptide of 120,000 daltons but can form higher molecular weight aggregates. The chromophore is thought to be a linear tetrapyrrole with some evidence suggesting that there is one chromophore per monomer (Tobin & Briggs, 1973). The proposed chromophore structures, their postulated linkages to the protein and a possible photo-isomerization mechanism are shown in Fig. 14.2 (Rüdiger, 1972).
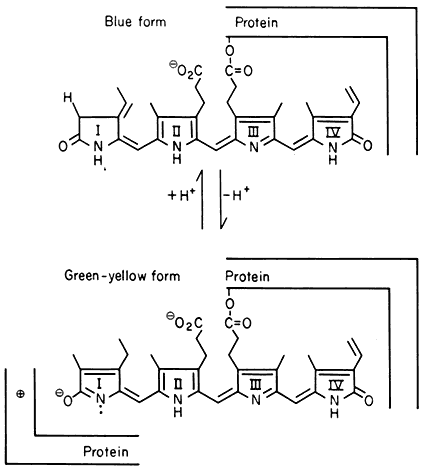
Figure 14.2
Proposed structure for the phytochrome chromophore, its linkage to the
protein and possible phototransformation mechanism (after Rüdiger, 1972).
The 'blue' form is thought to correspond to P r and the 'green-yellow' form to Pfr.
Information on differences in the molecular organization of the Pr and Pfr species has been sought in the hope that this might provide some insight into potential reaction mechanisms. Differences in the chromophore environment are evident from the absorption (Fig. 14.1), circular dichroism and optical rotatory dispersion spectra in the visible region (Kroes, 1970). Changes in protein conformation during photoconversion are also implied from low temperature and freeze-dry studies (Spruit & Kendrick, 1973; Kendrick, 1974). However, the post-conversion differences in the Pr and Pfr protein configurations are apparently only quite small as revealed by a variety of spectral and chemical methods (Briggs & Rice, 1972). Differences in surface residues are suggested by the ultraviolet difference spectrum (Tobin & Briggs, 1973); the differential reactivity of Pr and Pfr toward glutaraldehyde (Roux, 1972), and N -ethy maleimide (Gardner et al., 1974); and the differential electrostatic binding of Pr and Pfr to ribosomal material in plant extracts (Quail, 1975b).
14.3.2—
Photoconversion Reactions
Kinetic analysis following flash excitation of phytochrome indicates that the forward reaction (Pr ®Pfr ) requires several seconds to complete, whereas the reverse transformation (Pfr ®Pr ) is apparently complete by 20 to 30 msec
(Linschitz et al., 1966; Linschitz & Kasche, 1967). Several intermediates on separate pathways for the forward and reverse reactions have been characterized spectroscopically (Kendrick & Spruit, 1973). The first photochemical intermediate on both pathways appears to result from isomerization of the chromophore only with no change in protein structure. Subsequent dark relaxations apparently involve conformational changes in the protein moeity as well as further chromophore re-arrangements. The actual mechanism involved in chromophore photo-isomerization is uncertain, although tautomerization of the pyrrole group (Fig. 14.2) plus a cross-exhange of protons between chromophore and protein is a currently favoured hypothesis (Lhoste, 1972).
The phototransformation of a static population of phytochrome molecules can be described by the expression (Butler, 1972):
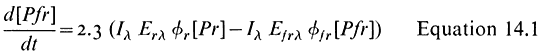
where l = wavelength; Il = intensity; t = duration of irradiation; Erl and Efrl = the extinction coefficients at l for Pr and Pfr respectively; fr and ffr = the quantum yields for Pr and Pfr respectively. At t =¥ a photo-equalibrium will be established and the pigment will oscillate ('cyc'e') between the two forms at a rate which is a function of the total absorption of the two species. The ratio of Pfr to Pr will remain constant. In the absence of any net loss or gain of phytochrome molecules in the population (a 'closed' system), this ratio will be wavelength dependent but irradiance independent according to the formula:

In the living cell, however, synthesis and destruction of phytochrome (see 14.3.3.2 below) must be taken into account. This transforms the system into an 'open' one where net loss or gain of pigment molecules can and do occur. For short term irradiations (about 5 minutes), where photo-equilibrium is rapidly established, no significant change in total phytochrome (Ptot ) occurs and Eq. 14.2 is a good first approximation. Under these conditions the [Pfr ]:[Pr ] ratio in the cell will be irradiance independent but wavelength dependent. Under long term continuous irradiations, however, synthesis and destruction become significant parameters with the result that the ratio [Pfr ]:[Pr ] becomes irradiance, as well as wavelength, dependent (Schäfer & Mohr, 1974; Schäfer, 1975).
Experimentally the kinetics of phototransformation have been shown to be first order both in vivo (Schmidt et al., 1973) and in vitro (Butler, 1961). Likewise, both the rates of photoconversion (Fig. 14.3) and the short term photosteady state ratio of Pr to Pfr (Fig. 14.4) have been demonstrated to be wavelength dependent in a manner consistent with the measured absorption spectra (Butler et al., 1964; Hanke et al., 1969).
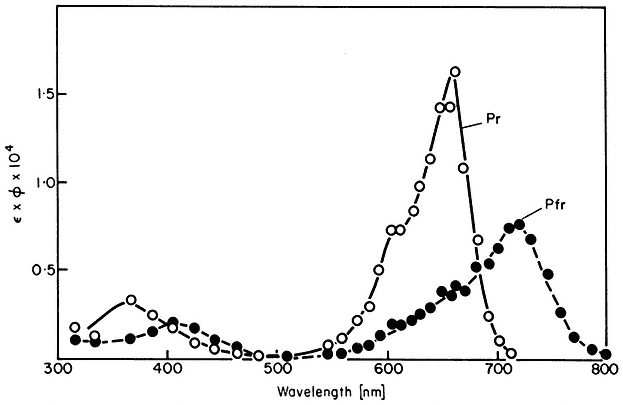
Figure 14.3
Action spectra of photochemical transformations of P r and Pfr in
solution. The extinction coefficient Î is in litre mol1 cm–1 and the
quantum yield f is in mol Einstein–1 (after Butler et al., 1964).
Note that the rate and extent of photoconversion are dependent on the wavelength, irradiance and time of irradiation below photo-equilibrium (Eq. 14.1). The product of time and irradiance determines the total number of quanta or the light 'dose' administered to the system. The wavelength determines how
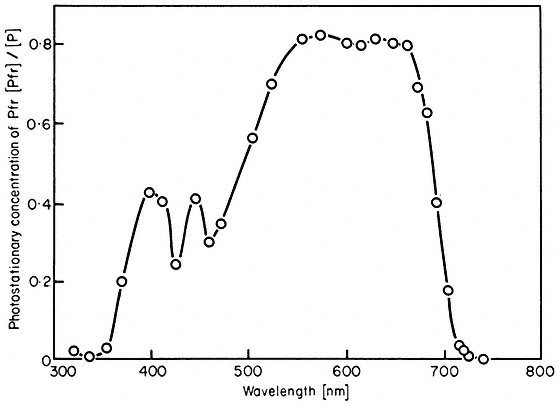
Figure 14.4
Proportion of phytochrome in the Pfr form at photoequilibrium
in vivo (Sinapis hooks) as a function of wavelength
(after K.M. Hartmann and C.J.P. Spruit in Hanke et al., 1969).
efficiently the incident quanta are absorbed by the pigment (Fig. 14.1). Note also that total photoconversion of Pr to Pfr is not possible (Fig. 14.4). The maximum attainable is about 80% Pfr in the red region of the spectrum. This is because there is no wavelength where Pr absorbs and Pfr does not (Fig. 14.1). Conversely, more than 97% of the phytochrome can be converted to the Pr form by wavelengths longer than 737 nm (Hartmann & Cohnen Unser, 1973) as the Pfr absorbance exceeds that of Pr in that region of the spectrum. The photosteady state ratio of Pfr/Pr can be conveniently manipulated with monochromatic light, particularly between 660 and 750 nm (Fig. 14.4). This procedure has been used to advantage in several physiological experiments.
For many years the only quantitative assay for phytochrome has been the spectrophotometric measurement of its photoreversible absorbance changes (Fig. 14.1) (Butler et al., 1959; Spruit, 1972). This procedure can measure phytochrome both in vivo and in vitro. However, it is unsuitable for use with green tissue and provides no index of the integrity of large regions of the protein moeity. The recent successful immunocytochemical detection of phytochrome now provides a second assay for the pigment (Coleman & Pratt, 1974).
14.3.3—
Dark Reactions
'Dark reversion' and 'destruction' are the so-called dark reactions of phytochrome. Neither the molecular bases nor the physiological significance of these processes is well understood.
14.3.3.1—
Dark Reversion
Pr is thermodynamically stable and can only be converted to Pfr by light (Lhoste, 1972). Pfr, in contrast, is metastable and can therefore revert thermally to Pr in the dark.
In vivo, dark reversion occurs in most dicotyledons but not monocotyledons, whereas phytochrome from both sources reverts in vitro (Frankland, 1972; Briggs & Rice, 1972). The process in vivo appears to be first order and rapid. In several plants reversion is complete within 30 minutes at 20ºC although only 15% to 20% of the Pfr molecules are involved. The remainder continue to undergo 'destruction' for a considerable period after reversion has ceased. Separate 'reversion' and 'destruction' pools of Pfr have been postulated to account for this apparent anomaly (Schäfer & Schmidt, 1974). Discontinuities in Arrhenius plots of the extent of reversion suggest that the process might be membrane associated (Schäfer & Schmidt, 1974).
14.3.3.2—
Synthesis and Destruction
Dry seeds contain phytochrome (Spruit & Mancinelli, 1969). Rapid, early increases in the photometrically detectable pigment (Ptot ) during imbibition are apparently due to rehydration of the molecule rather than synthesis (Tobin et al.,
1973). The pigment can be stored as either Pr or Pfr and rehydrated in the stored form (Vidaver & Hsiao, 1972). This suggests an explanation for the appearance of Pfr in the dark, sometimes observed in seeds (Rollin, 1972). Further increases in Ptot during the growth of etiolated seedlings result from de novo synthesis of new molecules in the Pr form (Quail et al., 1973b). The pigment accumulates to high levels in the dark ultimately reaching a plateau (Fig. 14.5).
'Destruction' is the disappearance of photometrically detectable Pfr without the concomitant appearance of equimolar quantities of Pr (Frankland, 1972). This decrease in photoactivity is paralleled by a loss of immunologically detectable phytochrome (Coleman & Pratt, 1974). Since no recycling of the protein moeity occurs, 'destruction' would appear to be a genuine degradative process (Quail et al., 1973b). Some evidence suggests that this process may be enzymatic (Kidd & Pratt, 1973). Destruction is temperature dependent, but the absence of discontinuities in Arrhenius plots suggests that in contrast to dark reversion, it is not membrane associated (Schäfer & Schmidt, 1974). Destruction is observed in both monocotyledons and dicotyledons with half-times ranging from 20 minutes to 4 hours (Frankland, 1972; Schäfer et al., 1973; Kidd & Pratt, 1973).
Destruction of Pfr occurs both in the dark following brief irradiations and in continuous light. In the dark, destruction rapidly removes all unreverted Pfr leading to a short-term decline in Ptot. The reduced pigment levels are replenished, however, as the dark period proceeds by de novo synthesis of new Pr molecules (Quail et al., 1973b). In continuous light, initially high Ptot levels decline at a rate proportional to the photosteady-state Pfr concentration (Frankland, 1972) ultimately reaching a new plateau (Fig. 14.5). This new
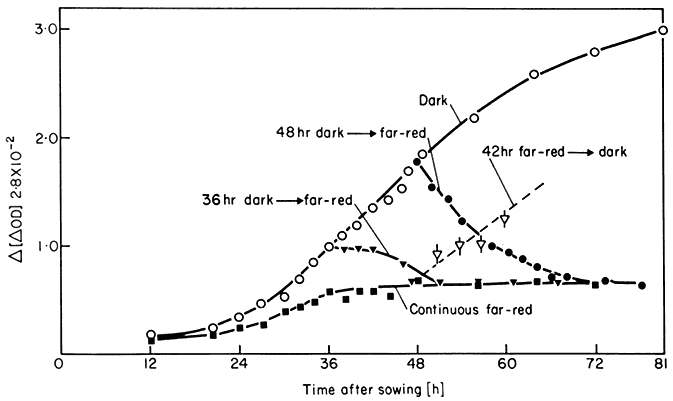
Figure 14.5
Total phytochrome levels in Sinapis cotyledons as function of time in the dark
(o); continuous far-red (


48 h dark ® far-red (

plateau represents a steady-state equilibrium between synthesis and degradation (Schäfer et al., 1972; Quail et al., 1973b). Pr synthesis appears to be a continuous zero order process, itself unaffected by light. The Ptot level is then regulated against this background by the disparate first order degradation rate constants for Pr and Pfr.
The plateau level of Ptot established at the steady-state by prolonged irradiations is a function of both wavelength and irradiance (Schäfer & Mohr, 1974; Schäfer, 1975). The Pfr level, in direct contrast however, is independent of both irradiance and wavelength under these conditions. This has the extremely important consequence that, whereas the ratio of [Pfr ]:[Ptot ] will change depending on wavelength and irradiance, the absolute level of Pfr will be the same under all continuous irradiation conditions once the steady-state has been established. This has important implications for the interpretation of the effects of prolonged irradiations.
14.3.3.3—
Physiological Significance of Dark Reactions
The dark reactions provide a mechanism for the light independent removal of Pfr and thereby the potential for a dark period to reverse light-induced responses which require the sustained presence of the effector. Furthermore, as the disappearance of Pfr is a time dependent process the system has the potential for measuring time. In principle, therefore, phytochrome should enable the plant to distinguish between light and dark and to time the dark period. Initially it was thought that this so-called 'hour-glass' principle was the basis for phytochrome-controlled photoperiodism (see Vince, 1972). Supporting evidence is scant, however, and this theory has now fallen into disrepute. Some systems do, nevertheless, respond to the timed disappearance of Pfr in the dark. An example is accumulation of the enzyme lipoxygenase (Oelse-Karrow & Mohr, 1973).
During prolonged irradiations, synthesis and destruction appear to have the additional role of maintaining a constant absolute level of Pfr irrespective of wavelength (Schäfer, 1975). This implies that Pfr per se is not the effector of high irradiance responses (see 14.4.1.2).
14.3.4—
Localization
The distribution of phytochrome is highly specific at the tissue and cellular levels as determined photometrically (Briggs & Siegelman, 1965) and immunocytochemically (Pratt & Coleman, 1971; 1974). In dicotyledons, the highest pigment levels are in the apical regions. In etiolated grass shoots, large quantities occur in parenchyma cells near the tip of the coleoptile and in the rapidly differentiating tissue near the shoot apex. High levels of the pigment are also found in root cap cells.
Attempts to establish the subcellular localization of phytochrome fall into two categories: (a) measurement of phytochrome-induced responses having a
spatial or vectorial component from which the photoreceptor location can be inferred; (b) direct measurements of the pigment itself, either in situ or in subcellular fractions.
The pattern of chloroplast movement in the alga Mougeotia in response to polarized red and far-red microbeams has been taken as evidence that phytochrome located and orienled on or near the plasmalemma controls this response (Haupt, 1972b). The directional growth of the germ tubes of Dryopteris in polarized light has been interpreted similarly (Etzold, 1965). The change in ion flux associated with phytochrome mediated leaflet movement (Satter & Galston, 1973), root tip adhesion to glass surfaces (Tanada, 1968), and changes in bioelectric potentials (Newman & Briggs, 1972) are also indicative of phytochromecontrolled changes in surface properties but not necessarily that the pigment is a permanent membrane component. A report of phytochrome-controlled development of isolated etioplasts in vitro (Wellburn & Wellburn, 1973) implies the presence of functionally active pigment in or on the organelles. More recently a rapid, phytochrome-mediated change in the level of gibberellin extractable from an etioplast-rich fraction in response to in vitro irradiations has been demonstrated (Evans, 1975; Evans & Smith, 1976a; Cooke & Saunders, 1975). A red/far-red reversible reduction of NADP in vitro in response to irradiation of a mitochondria-rich fraction has also been reported (Manabe & Furuya, 1974). Furthermore, phytochrome has been detected spectrophotometrically in both etioplast- (Evans & Smith, 1975) and mitochondria-rich fractions (Manabe & Furuya, 1974).
Both spectrophotometric and immunological techniques have been used for direct, in situ measurements of the intracellular distribution of phytochrome. The presence of the pigment in the nucleus has been claimed on the basis of microspectrophotometric scans of cells (Galston, 1968) but these data have been challenged (Kendrick & Spruit, 1972; Tobin et al., 1973). The cytochemical visualization of phytochrome antibody in non-irradiated tissue sections indicates a general distribution of the photoreceptor throughout the cytoplasm in addition to an association with nuclei and plastids (Pratt & Coleman, 1971). Brief red irradiation prior to fixation causes the pigment in some tissues to concentrate in discrete, as yet unidentified regions of the cytoplasm (Mackenzie et al., 1974). Non-saturating irradiations of maize coleoptile segments with red and far-red light polarized normal to the longitudinal axis were found to photoconvert about 20% more phytochrome than when polarized parallel to this axis (Marmé & Schäfer, 1972). This was interpreted as indicating that phytochrome is located and oriented in the plasmalemma. In considering the locational and orientational rigidity of phytochrome implied from polarized light studies the known rapid and highly fluid lateral and rotational diffusion of other membrane proteins should be borne in mind (Cone, 1972; Singer, 1974).
Cell fractionation procedures have also been used in attempts to localize phytochrome in subcellular components. The well-known precipitation of the pigment protein at low pHs (
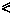
has been overlooked in some studies leading to claims of associations of phytochrome with mitochondria (Gordon, 1961) and plasmalemma (Marmé et al., 1971). Little of the pigment (< 10%) sediments from homogenates of nonirradiated tissue at neutral pH at forces up to 144,000 × g (Rubinstein et al., 1969; Siegelman & Butler, 1965). Red irradiation prior to extraction, however, substantially enhances the level of phytochrome subsequently associated with pelletable material (Quail et al., 1973a). Irradiation of extracts from dark grown material has a similar effect (Marmé et al., 1973). Initially claims were made of the isolation of a phytochrome-containing membrane fraction that could be reversibly 'solubilized' by withdrawal of Mg2+ (Marmé et al., 1973; 1974). More recently, however, the pigment in this fraction has been shown to be associated with degraded ribonucleoprotein (RNP) material, probably of ribosomal origin (Quail, 1975b). This association apparently results from the preferential electrostatic adsorption of Pfr onto ribosomal material—either free or membrane-bound in the endoplasmic reticulum (Williamson et al., 1975). Whether such an association is artefactual or biologically meaningful is yet to be established. A recent promising variation on this approach is the use of glutaraldehyde in an attempt to immobilize the pigment in the cell prior to extraction (Yu, 1975.)
The existence of meaningful phytochrome-membrane interactions are by no means excluded by the above findings. It has been shown, for example, that phytochrome can mediate photoreversible conductance changes in artificial lipid membranes (Roux & Yguerabide, 1973). The suggestion that phytochrome might function as a stereospecific protein ligand capable of interaction with cellular membranes has been made (Quail & Schäfer, 1974; Boisard et al., 1974).
14.4—
Phytochrome Physiology
The biological responses attributed to phytochrome can be usefully characterized in terms of three different but interrelated sets of criteria: (a) the nature of the involvement of light in the inductive process; (b) the temporal expression of the induced response; and (c) the type of cellular or developmental process affected.
14.4.1—
Induction-Reversion and High Irradiance Responses
Two types of light-controlled phenomena have been attributed to phytochrome—the so-called 'induction-reversion' and 'high irradiance' (HIR) responses (Mohr, 1972). This terminology arises from the irradiation conditions under which the responses are observed and suggests a fundamental difference in the manner in which the phytochrome molecule transmits the light signal to the cell in each case. It does not necessarily reflect an intrinsic property of the actual, biological parameter being monitored. Some parameters display both modes of response, others only one.
14.4.1.1—
Induction-Reversion Responses
These are the classical phytochrome responses (Borthwick, 1972). A change in the biological parameter being monitored is induced by a brief irradiation of low intensity red light and reversed by a subsequent far-red pulse. The accumulation of anthocyanin in Sinapis illustrates this point (Fig. 14.6). Another well known example is lettuce seed germination. This is repeatedly photoreversible for up to 100 alternate red and far-red irradiations (Borthwick, 1972).
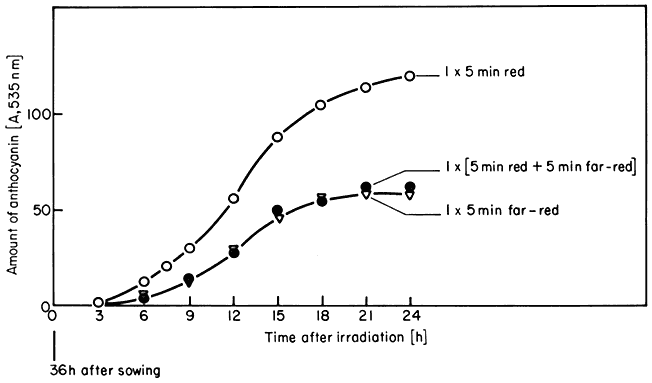
Figure 14.6
Accumulation of anthocyanin in Sinapis in the dark following irradiation
treatments at time zero with 5 min red (o), 5 min far-red (Ñ ), or 5 min
red followed immediately by 5 min far-red (

Mohr et al., 1971).
This simple red/far-red photoreversibility forms the basis of the concept that Pfr is the biologically active form of the pigment whereas Pr is inactive. Attempts to quantify the relationship between the number of Pfr molecules formed and the magnitude of the induced response have been both indirect and direct.
Indirect correlations are based on the premise that observed increases in the magnitude of the response with increasing light dose are a function of the degree of photoconversion of Pr to Pfr, i.e. the more quanta, the more Pr is converted to Pfr and therefore the greater is the response. The increase in anthocyanin in response to increasing doses of red light (Table 14.1) illustrates this point (Lange et al., 1971). The so-called law of reciprocity (irradiance × time = constant) must hold for the light doses used for this interpretation to be valid (see 14.3.2). This establishes that the magnitude of the response is directly proportional to the total number of incident quanta regardless of the time or irradiance of the
irradiation providing those quanta (Table 14.1). The effects of irradiance level during the brief irradiations used in induction-reversion experiments are thus attributed entirely to the degree of photoconversion.
Biological action spectra are an extension of this principle (Fig. 14.7). The magnitude of the response at different wave-lengths is interpreted to be a function of the relative effectiveness of the quanta at those wavelengths in the phytochrome photoconversion process. The close agreement between the action spectra of several biological responses on the one hand (Fig. 14.7) and those of the phototransformation reactions of the isolated pigment on the other (Fig. 14.3) lends strong support to this notion (Borthwick, 1972; Shropshire, 1972). In these cases a seemingly good correlation exists between Pfr level and response magnitude.
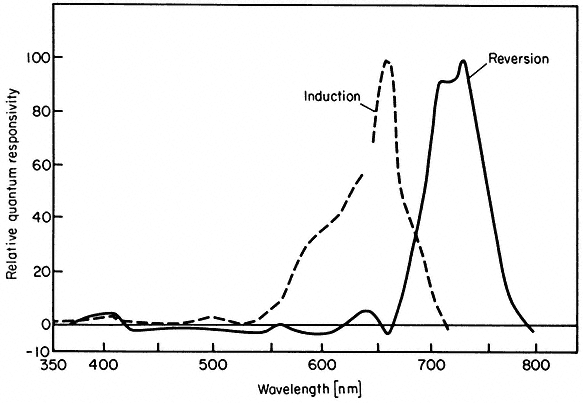
Figure 14.7
Action spectra for induction and reversion of plumular hook opening in bean seedlings
(after Withrow et al., 1957).
In contrast, however, the majority of rigorous attempts to demonstrate a direct quantitative correlation between the photometrically detectable Pfr level and the relevant biological response in the same system have been unsuccessful (Hillman, 1972). The reasons for this are not understood. An apparent exception is lipoxygenase levels in Sinapis (Oelze-Karrow & Mohr, 1973).
Increases in response with increasing doses of quanta must eventually saturate. If the level of Pfr is rate-limiting the photoresponse will saturate when the photoconversion process is saturated i.e. when photoequilibrium is reached. If the response system itself is rate-limiting the response may saturate well before photoequilibrium. Examples of both extremes have been observed (Hillman, 1967). Light doses which saturate the photoconversion of Pr to Pfr do not appear to saturate the inhibition of mesocotyl lengthening in Avena (Loecher, 1966), nor the accumulation of anthocyanin in Sinapis (Drumm & Mohr, 1974). In contrast, inhibition of lipoxygenase accumulation is saturated by very low (< 3%) Pfr levels (Mohr, 1972). Other photoresponses fall between these extremes with many being saturated at less than 80% Pfr (Hillman, 1967). In addition, whereas many parameters, such as anthocyanin formation, show a graded response, others, such as lipoxygenase accumulation, respond in an all-or-none fashion to changes in Pfr level, suggesting some form of cooperative, threshold mechanism (Oelze-Karrow & Mohr, 1973).
Implicit in the far-red reversibility of an induced response is that Pfr can act in the dark. Light is strictly a trigger. The magnitude and multiplicity of the responses indicate an extensive amplification mechanism. Unlike photosynthesis where light energy is converted stoichiometrically with quantum yields of less than 1.0, the low irradiances which actuate these phytochrome responses lead to final quantum yields well in excess of unity (Galston, 1974).
14.4.1.2—
High Irradiance Responses (HIR)
If Pfr is, as postulated, the active, effector, it is clear that tripartite correlations between light dose, Pfr level and response magnitude will only be expected for irradiations terminated prior to photoequilibrium. Once photoequilibrium has been established, Pfr le vels are no longer irradiance dependent whether for short term (Eq. 14.2) or long term (Schäfer, 1975) irradiations. No further increase in the response should result therefore from further increases in irradiance after photoequilibrium.
Some parameters do, however, show a strong irradiance dependence at photoequilibrium. This effect is termed the high irradiance response (HIR) (Mohr, 1969). Such responses are observed with continuous irradiation where a phytochrome photosteady-state is rapidly established and maintained for prolonged periods. The accumulation of anthocyanin in continuous far-red light illustrates this point (Fig. 14.8) (Lange et al., 1971). The rate of accumulation is a function of the irradiance. This effect is only maintained, however, as long as the irradiation continues. The irradiance-enhanced response rate reverts rapidly
to that of the dark controls when irradiation ceases, and resumes again upon further irradiation (Fig. 14.9). Reciprocity is not demonstrable for the HIR and, in some cases e.g. lettuce hypocotyl lengthening, the variable itself does not exhibit the classical red/far-red reversible response (Hartmann, 1966). Anthocyanin accumulation, in contrast exhibits both modes of response (Figs. 14.6 and 14.8).
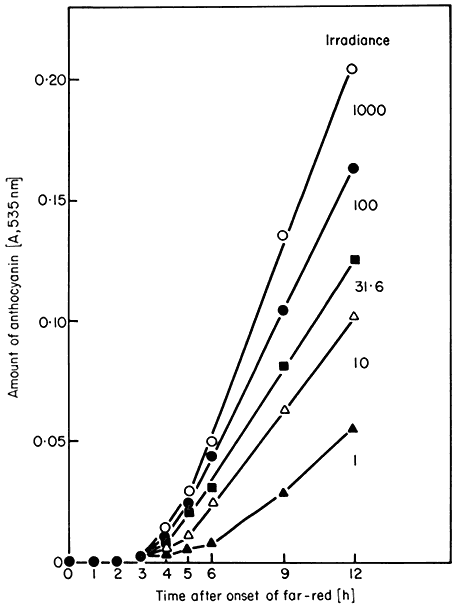
Figure 14.8
Accumulation of anthocyanin in Sinapis under high irradiance
conditions. Seedlings were held in continuous far-red light of
varying irradiance. All irradiances are expressed relative
to the arbitrary value of 1,000 (= 350 µW. cm–2 )
(after Lange et al., 1971).
Action spectra of HIR have peaks in the blue and far-red (Borthwick et al., 1969; Mohr, 1969). The most extensively studied response is that of the inhibition of lettuce hypocotyl lengthening (Hartmann, 1967; see also Fig. 14.10). The seedlings were irradiated continuously for 18 hours with monochromatic light of different wavelengths and irradiances. As with other HIR there is no coincidence of the observed spectrum with the absorption maxima of either Pr or Pfr.
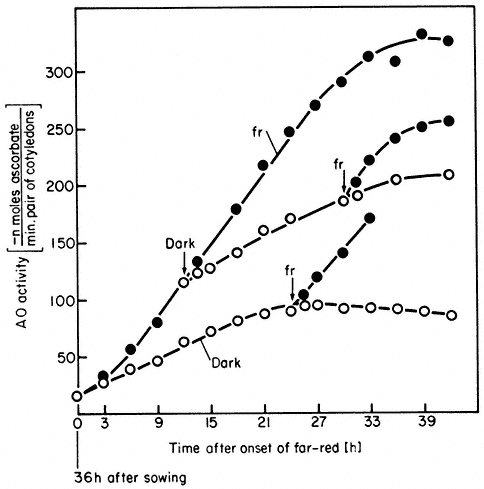
Figure 14.9
Accumulation of ascorbate oxidase activity in Sinapis under
high irradiance conditions. Seedlings were either retained in
the dark (o) or continuous far-red
seedlings from far-red to the dark or vice versa at
various times is indicated by the arrows (̄ )
(after Drumm et al., 1972).
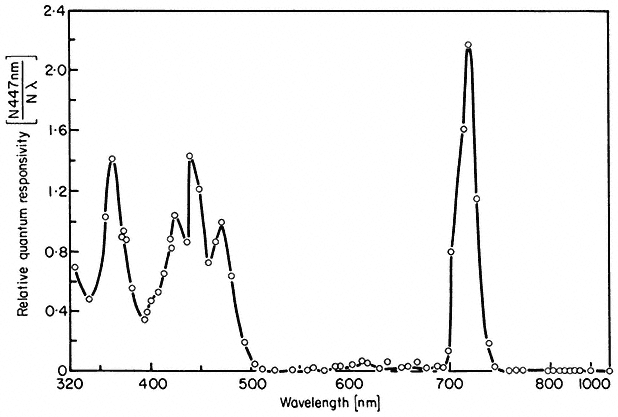
Figure 14.10
Action spectrum for inhibition of lettuce hypocotyl
lengthening under continuous irradiation
(after Hartmann, 1967).
The conclusion that phytochrome mediates the high irradiance effects derives from other data using the same plant system (Hartmann, 1966). Prolonged irradiations with two wavelengths which were relatively ineffective when given separately (658 nm and 766 nm) were highly effective when given simultaneously. The maximum effect with these and other wavelength pairs always occurred where the photosteady-state Pfr concentration was about 3%. This agrees well with the peak of activity observed at 720 nm with single wavelength monochromatic light (Fig. 14.10) also known to establish about 3% Pfr at photoequilibrium (Fig. 14.4). Furthermore, the effectiveness of a single wavelength irradiation at 717 nm could be nullified by simultaneous irradiations of either 658 nm or 759 nm. These wavelengths would shift the photoequilibrium away from 3% Pfr towards higher or lower values respectively.
The question remains, however, as to how both the irradiance and wavelength dependency of the HIR can be explained in terms of phytochrome. The response is unlikely to be a function of the absolute Pfr level as this is irradiance and wavelength independent with prolonged irradiations (Schäfer, 1975). The rate at which the pigment molecule oscillates between the two forms at photoequilibrium is, on the other hand, strongly irradiance and wavelength dependent. Accordingly, the HIR has been rationalized to be some function of the cycling rate of phytochrome. The effector molecule has been postulated to be some 'excited form of Pfr', denoted Pfr* (Schopfer & Mohr, 1972). No direct evidence for such a species is available, however, and its purported action has been questioned on the basis of dual wavelength experiments (Hartmann & Cohnen Unser, 1973). High levels of phytochrome intermediates are maintained at high irradiances (Kendrick & Spruit, 1973a) but the possibility that these are HIR effectors is unlikely because of the nature of the action spectra (Hartmann, 1966).
14.4.1.3—
A Unitary Model
Schäfer (1975) has recently developed a single, formalistic model which can theoretically account for both induction-reversion and HIR responses, whether graded or cooperative, in terms of phytochrome. The pigment is postulated to be a bimodal ligand with the forms Pr and Pfr which interact with receptor sites also having dual forms, X and X':
where °ks and kd are the rate constants for synthesis and destruction of phytochrome respectively; k1 and k2 rate constants for the photoconversion reactions; and k3 and k4 rate constants for the X®X' and X'®X transitions.
The irradiance and wavelength dependencies of the HIR response then become explicable in terms of the flux rates through the cycle under continuous irradiation. The basic conclusions reached are that PfrX' is the effector element in induction-reversion responses, whereas PfrX or the flux k3 . PfrX is the effector element in HIR. The molecular nature of X and the mechanism of action of the effector elements are, however, unresolved questions.
14.4.1.4—
Modes of Light Signal Transmission
Given that phytochrome mediates both induction-reversion and HIR, it is clear that the photoreceptor utilizes the incoming light signal differently in each case. Whereas induction-reversion responses are induced by a red pulse and can develop in the subsequent dark period, HIR require a continuous light energy input to sustain the response. Increases in irradiance lead to increases in response magnitude in both cases but the effect is interpreted differently for each. For induction-reversion responses the irradiance effect is only observed prior to photoequilibrium; is interchangeable with time of irradiation (reciprocity holds); and is interpreted as reflecting the effectiveness of the total light dose in determining the degree of photoconversion of Pr to Pfr. Pfr is considered the effector molecule and some form of Pfr -response stoichiometry is expected. For HIR, on the other hand, the irradiance effect is observed at photoequilibrium; is not interchangeable with time of irradiation (reciprocity does not hold); and is interpreted as being some function of the phytochrome cycling rate. The precise effector molecule or process is uncertain. For induction-reversion responses light is viewed simply as a trigger and as having no further direct role in the inductive function of Pfr. The light signal is 'stored' in the Pfr form for subsequent utilization. HIR, in contrast, require a sustained, direct interaction of the photoreceptor with the incident excitation energy. This indicates that these effects are light-driven as distinct from being light-triggered. Light appears to have a direct role in the inductive function of the pigment, the energy input being rapidly dissipated.
The suggestion that photosynthesis or cyclic photophosphorylation might in some way be responsible for HIR has been advanced but several pieces of evidence argue against this (Mohr, 1972). Speculative suggestions that phytochrome might function as a photocoupler (Quail, 1975a) or a specific, lightdriven permease (Smith, 1970) in the HIR have also been advanced but not substantiated.
14.4.2—
Response Kinetics
Phytochrome is considered to trigger, in some way, a chain of events leading sooner or later to a measurable biological response. The initial triggering of those processes necessary for the development of the response can be termed phytochrome 'action'; and the appearance of a measurable change in the
parameter being monitored can be called response 'expression'. This leads to the recognition of three categories of phenomena: (a) rapid action/rapid expression; (b) rapid action/delayed expression; and (c) delayed action/delayed expression responses. 'Rapid' here arbitrarily means
14.4.2.1—
Rapid Action/Rapid Expression Responses
As phytochrome action must either coincide with or precede expression, rapid action can be implied from the kinetics of the expression alone in these cases. A red/far-red reversible change in electric potential in Avena coleoptiles within 15 seconds of the start of irradiation is the most rapid phytochrome-mediated phenomenon thus far reported (Newman & Briggs, 1972). Similar changes have been observed in the biolelectric potential of mung bean root tips exposed to successive red and far-red irradiations (Jaffe, 1968; Racusen & Miller, 1972). Such changes had earlier been inferred from the red/far-red reversible adhesion of root tips to negatively charged glass surfaces (Tanada, 1968). Both responses are detectable within 30 seconds of the start of irradiation (Fig. 14.11). These changes in surface charge are interpreted to indicate changes in the plasmalemma. Fluorescent probe studies support this notion (Racusen, 1973). Red light also induces H+ efflux from root tips (Yunghans & Jaffe, 1972).
Phytochrome regulates leaflet movement in Mimosa, Albizia and Samanea (Satter & Galston, 1975). This movement is accompanied by an energy-dependent transfer of K+ ions between the ventral and dorsal motor cells of the pulvinus. Both effects are detectable 10 minutes after red or far-red irradiations. The changes in K+ are correlated with changes in transmembrane potential (Racusen & Satter, 1974). Furthermore, red/far-red regulated changes in surface charge similar to those of root tips are detectable 30 to 120 seconds after irradiation. These effects are also strongly indicative of phytochrome-mediated changes in plasmamembrane properties.
Phytochrome-regulated changes in the rate of plasmolysis have been detected in Mougeotia within 6 minutes of the onset of red irradiations (Wiesenseel & Smeibidl, 1973). A change in plastid orientation is also evident in this alga in less than 10 minutes after a red pulse (Haupt, 1972a). The latter effect can only be fully reversed by far-red during the first minute after red light. Thus potentiation of the response has begun within 1 minute of photoconversion. Available data indicate that the effective phytochrome is located on or near the plasmalemma. Contractile fibrils appear to be responsible for the chloroplast movement (Schönbohm, 1973).
Red light (15 seconds) induces an increase in growth rate in coleoptile tips within 60 seconds of the start of irradiation (Weintraub & Lawson, 1972). The effect is partially reversed if far-red immediately follows the red. Inhibitors of transcription and translation are without effect. As the cell must regulate wall extension through the plasmalemma, this response might also represent a
Figure 14.11
Kinetics of (A) root tip adhesion to a negatively charged glass
surface and (B) the development of a biolectric potential across
the root tip in response to irradiation with red or far-red light
(after Jaffe, 1968).
phytochrome-mediated membrane change. Rapid changes in ATP levels, induced by red and, in some cases, partially reversed by far-red, have been reported but no consistent pattern is obvious (Sandmeier & Ivart, 1972; Yunghans & Jaffe, 1972; White & Pike, 1974).
Rapid transitory increases in hormore levels in response to red light have been known for some time (Reid et al., 1968; Beevers et al., 1970; van Staden & Wareing, 1972). Unequivocal evidence of far-red reversibility has generally been lacking in the past, however, often because of poor experimental design. Recently, the level of gibberellin activity extractable from etioplast-rich preparations from grass leaves has been shown to increase 3-fold within 5 minutes of the termination of a 5 minute red irradiation of the isolated fraction (Evans, 1975; Evans & Smith, 1976a). This effect is reversed by far-red light given immediately after the red. It is postulated that phytochrome causes the movement of gibberellin across the etioplast envelope into the ambient medium.
These observations, and those made with mitochondria-rich fractions (Manabe & Furuya, 1974), are rapid, of potential physiological significance, and suggestive of in vitro phytochrome-mediated changes in the functional properties of membranes in the isolated fractions.
Lipoxygenase accumulation in Sinapis responds to changes in Pfr level in less than 5 minutes under appropriate conditions (Oelze-Karrow & Mohr, 1973). It has been concluded that the level of the enzyme in the cotyledons is controlled by phytochrome in the hypocotyl hook, through a highly cooperative threshold mechanism that responds to, and is saturated by, Pfr levels of 1–2% of the total pigment (Oelze-Karrow & Mohr, 1974). Other rapid, inter-organ transfer of phytochrome signals have also been reported (de Greef & Caubergs, 1973). Some form of biophysical transmission system has been postulated, with the membrane continuum of the plasmodesmata as a suggested candidate (Oelze-Karrow & Mohr, 1973).
14.4.2.2—
Rapid Action/Delayed Expression Responses
Rapid phytochrome action in these cases is deduced from the rate at which the response escapes susceptibility to photoreversal by far-red light following an inductive red pulse. The actual response may not be expressed for days after the irradiation although the inevitability of its appearance has long since been irreversibly established. Pfr is said to have 'potentiated' the response (Borthwick, 1972). The escape from reversibility is viewed as the Pfr -triggered reaction chain having progressed beyond those steps directly under phytochrome confrol.
The effect of a red light pulse on flowering in Pharbitis is only partially reversed by far-red given 30 seconds after the start of the red irradiation (Fredericq, 1964). After 3 minutes, far-red no longer reverses the effect. Thus although the flowering response itself is not expressed for several days, it is potentiated within seconds by Pfr. Flowering in Chenopodium album and Kalanchoe behaves similarly. A synergism between phytochrome and gibberellin in lettuce seed germination (Bewley et al., 1968) and the de-etiolation response of Pisum (Haupt, 1972a) are also in this category.
14.4.2.3—
Delayed Action/Delayed Expression Responses
These responses show a lag from irradiation to expression but are readily reversed by far-red over relatively long periods in the dark after the red pulse. Gradual escape from reversibility can occur but is relatively slow. This is viewed as indicating that the continued presence of Pfr is required over a relatively long period in the dark to maximise expression. An important feature of these responses is that dark reversion and destruction are continually and often rapidly depleting the Pfr pool during this period.
The vast majority of recorded red/far-red reversible responses, too numerous to catalogue, are included in this category (see Mitrakos & Shropshire, 1972;
Mohr, 1972; Smith, 1975). Lettuce seed irradiated for I minute with red light germinates up to 100% 24 hours later. Far-red reverses this effect up to 12 hours after the red irradiation but with decreasing effectiveness. Anthocyanin formation in Sinapis has a lag of 3 hours from irradiation to the onset of accumulation (Fig. 14.6). During this time the effect becomes decreasingly susceptible to reversal by far-red but escape is never complete. The in vitro protein synthetic capacity of 80s ribosomes from corn is enhanced by 5 minutes of red light prior to harvest (Travis et al., 1974). This effect is detectable within 30 minutes of red irradiation and escapes far-red reversibility within an hour.
14.4.3—
Response Manifestations
Phytochrome has a regulatory role in all major phases of plant growth and development. The changes in cellular biochemistry and physiology which underlie this regulation are detectable at almost any chosen level. Some responses are biophysical in nature (membrane potential), others biochemical (enzyme levels); some require protein synthesis (leaf expansion), others do not (leaf movements); some involve cell division (seed germination), others only cell expansion (plumular hook opening); some are restricted to the irradiated cells (leaf movements), others result from transmissable stimuli (floral induction); some are dependent on phytochrome-endogenous rhythm interactions (flowering), others appear independent (seed germination) (Galston, 1974; Satter & Galston, 1975).
When the nature of the responses is coupled with their kinetic properties a pattern emerges. In general, the most rapid responses are surface or membrane-associated phenomena, often physico-chemical in nature and independent of RNA and protein synthesis. Other cellular processes such as changes in enzyme levels mostly respond more slowly.
14.5—
Mechanism of Action
The molecular mechanism of phytochrome action is the central issue in phytochrome research. The term 'primary reaction of phytochrome' has been used to describe the first reaction in which the physiologically active species becomes involved (Mohr, 1972). Assuming Pfr to be the active species this reaction has been formalized:
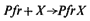
where X satisfies the logical necessity of a reaction partner regardless of its nature. The 'primary action' of phytochrome then is presumably the induction of a functional change in the reaction partner (X ®X' ). These are useful formalistic treatments but say nothing of the actual molecular mechanisms involved.
Any molecular hypothesis of phytochrome action must provide an explanation for the specificity, multiplicity and amplification of the induced responses, and attempt to reconcile the seemingly disparate modes of light signal transmission in induction-reversion and high irradiance responses. Specificity here refers to the observed capacity of different plants, organs, adjacent cells in the same tissue and even different processes in the same cell to respond differently to the same effector molecule (Mohr, 1972). All major hypotheses hold the view that this property resides in the fundamental programming of the target cells themselves rather than in the phytochrome molecule.
Multiplicity refers to the vast diversity in form, direction, magnitude and timing of response expression per se (see 14.4.3). This can be accounted for by a suitable amplification mechanism. Two major hypotheses of phytochrome action, based on this rationale, have been advanced in recent years: the differential gene activation hypothesis (Mohr, 1966) and the membrane permeability hypothesis (Hendricks & Borthwick, 1967). The possibilities that plant hormones (Black & Vlitos, 1972) or acetylcholine (Jaffe, 1970) might be primary mediators of phytochrome effects have also been investigated but convincing supporting data are sparse (Tanada, 1972).
The gene activation hypothesis in its broadest sense is trivial, as gross morphogenetic changes such as flowering are unlikely to occur without some modification of gene expression. In its strictest sense the hypothesis implies that the primary action of phytochrome is to alter genetic activity. This requires direct Pfr -genome interaction. Data relevant to this problem come mainly from measurements of phytochrome-induced changes in enzyme activities and related phenomena (Mohr, 1972).
There are two major criticisms of this hypothesis. First, while the data so far obtained are consistent with the general framework of the concept, they provide no evidence that Pfr interacts directly with the genome. Increased incorporation of [-H]-uridine into high molecular weight RNA in continuous far-red light has been reported and interpreted as phytochrome-enhanced transcription of rRNA cistrons (Thien & Schopfer, 1975). Phytochrome-mediated de novo synthesis of enzyme proteins has also been demonstrated (Attridge, 1974; Acton & Schopfer, 1974) but this does not establish that control is at the gene level. Indeed, insensitivity of responses such as triose phosphate dehydrogenase (Cerff, 1974) and ascorbate (Schopfer, 1967) accumulation towards actinomycin D might suggest otherwise in these cases. Evidence of phytochrome-mediated activation of precursors of phenylalanine ammonia-lyase has been advanced (Attridge et al., 1974) but this point is controversial (Acton & Shopfer, 1975).
The second criticism is that a growing list of responses appear unlikely to be explained by changes in gene expression. These are in general the rapid action/rapid expression responses. These are more rapid than would be expected of gene regulation and are often insensitive to inhibitors of transcription and translation (Satter & Galston, 1973). Furthermore, lipoxygenase, the only enzyme responding rapidly enough to be placed in this category, is now postu-
lated to be controlled by phytochrome in a separate organ (Oelze-Karrow & Mohr, 1974).
The membrane hypothesis postulates that phytochrome interacts directly with cellular membranes i.e. the primary action of Pfr is to modify membrane properties. Indirect support for this notion derives from the fact that the majority of the most rapid responses are either surface phenomena or can be rationalized in terms of membrane changes (see 14.4.2.1). Direct evidence of an association of phytochrome with an identifiable cellular membrane is yet to be advanced however (see 14.3.4) although Evans and Smith (1976b) have recently reported the presence of phytochrome in fractions enriched for etioplast envelope membranes.
A postulated membrane locale of phytochrome action permits the formulation of a unitary hypothesis to account for all phytochrome-mediated phenomena. Both rapid and long term effects can be rationalized to have emanated from the same primary event. A single, fundamental alteration in membrane properties, perhaps simultaneously in several cellular membranes, provides an immediate mechanism for the generation and amplification of a multiplicity of secondary effects (Changeux, 1969). Such effects might include changes in ion flux, activation of membrane bound enzymes, altered compartmentalization and soon. Any or all of these might lead directly or indirectly to altered gene expression. Moreover, a phytochrome-membrane association suggests a possible basis for the dual function of the pigment in induction reversion and HIR responses. The opportunity would exist for the molecule to function either as a reversible, stereospecific membrane effector, via photoinduced changes in its protein conformation; and/or as a photocoupler (Quail, 1975a) or light driven permease (Smith, 1970) via pigment cycling.
The speculative nature of such suggestions, however, is evidence of our ignorance in this area. Indeed the basic concept of only a single, primary phytochrome reaction has been challenged (Mohr, 1974). It has been suggested instead that a multiplicity of reaction partners and corresponding primary reactions must be postulated to account for both graded and cooperative responses in the same system (Mohr et al., 1971). This would circumvent the second criticism of the gene regulation hypothesis and obviate any potential conflict between the two major hypotheses. The molecular mechanism of phytochrome action is clearly an open question.
Further Reading
Briggs W.R. & Rice H.V. (1972) Phytochrome: Chemical and physical properties and mechanism of action. Ann. Rev. Plant Physiol.23, 293–34.
Galston A.W. (1974) Plant photobiology in the last half-century. Plant Physiol.54, 427–36.
Hillman W.S. (1967) The physiology of phytochrome. Ann. Rev. Plant Physiol.18 , 301–24.
Mitrakos K. & Shropshire W. JR. (1972) Phytochrome. Academic Press, London and New York.
Mohr H. (1972) Lectures on Photomorphogenesis. Springer-Verlag, Berlin.
Mohr H. (1974) Advances in phytochrome research. In Photochem. Photobiol. (in press).
Satter R.L. & Galston A.W. (1975) The physiological functions of phytochrome (in press).
Siegelman H.W. & Butler W.L. (1965) Properties of phytochrome. Ann. Rev. Plant Physiol. 16, 383–93.
Smith H. (1970) Phytochrome and photomorphogenesis in plants. Nature227, 665–8.
Smith H. (1975) Phytochrome and Photomorphogenesis. McGraw-Hill, U.K. Ltd., Maidenhead.